A survey on cellular RNA editing activity in response to Candida albicans infections
- PMID: 29363428
- PMCID: PMC5780849
- DOI: 10.1186/s12864-017-4374-2
A survey on cellular RNA editing activity in response to Candida albicans infections
Abstract
Background: Adenosine-to-Inosine (A-to-I) RNA editing is catalyzed by the adenosine deaminase acting on RNA (ADAR) family of enzymes, which induces alterations in mRNA sequence. It has been shown that A-to-I RNA editing events are of significance in the cell's innate immunity and cellular response to viral infections. However, whether RNA editing plays a role in cellular response to microorganism/fungi infection has not been determined. Candida albicans, one of the most prevalent human pathogenic fungi, usually act as a commensal on skin and superficial mucosal, but has been found to cause candidiasis in immunosuppression patients. Previously, we have revealed the up-regulation of A-to-I RNA editing activity in response to different types of influenza virus infections. The current work is designed to study the effect of microorganism/fungi infection on the activity of A-to-I RNA editing in infected hosts.
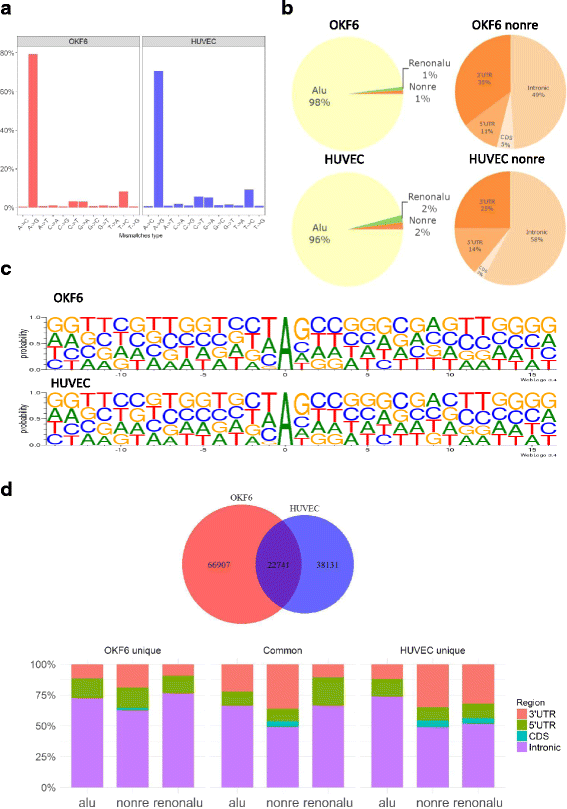
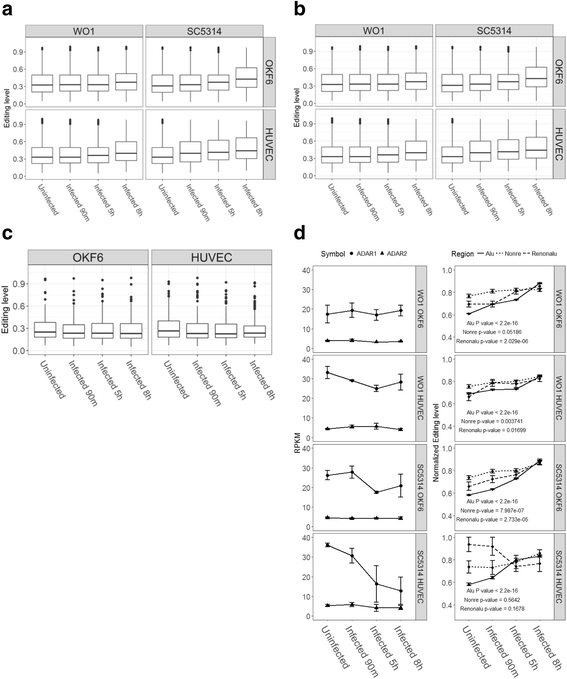
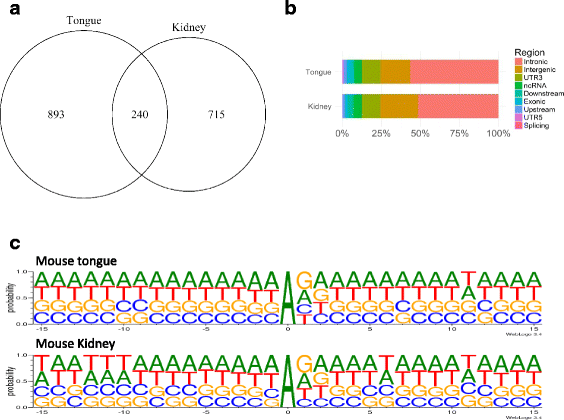
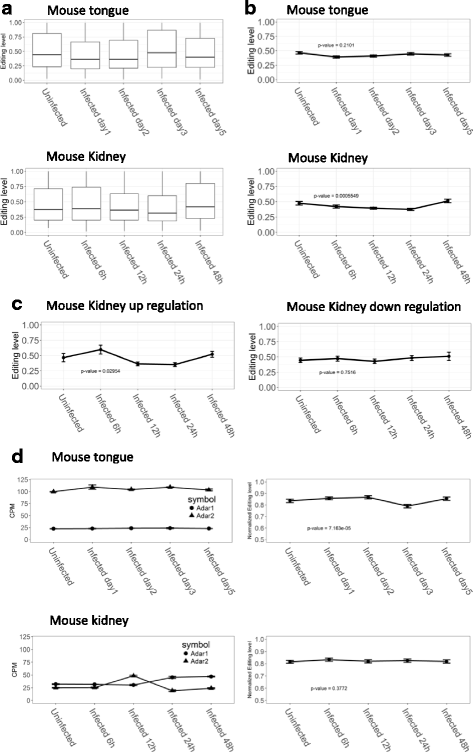
Results: We first detected and characterized the A-to-I RNA editing events in oral epithelial cells (OKF6) and primary human umbilical vein endothelial cells (HUVEC), under normal growth condition or with C. albicans infection. Eighty nine thousand six hundred forty eight and 60,872 A-to-I editing sites were detected in normal OKF6 and HUVEC cells, respectively. They were validated against the RNA editing databases, DARNED, RADAR, and REDIportal with 50, 80, and 80% success rates, respectively. While over 95% editing sites were detected in Alu regions, among the rest of the editing sites in non repetitive regions, the majority was located in introns and UTRs. The distributions of A-to-I editing activity and editing depth were analyzed during the course of C. albicans infection. While the normalized editing levels of common editing sites exhibited a significant increase, especially in Alu regions, no significant change in the expression of ADAR1 or ADAR2 was observed. Second, we performed further analysis on data from in vivo mouse study with C. albicans infection. One thousand one hundred thirty three and 955 A-to-I editing sites were identified in mouse tongue and kidney tissues, respectively. The number of A-to-I editing events was much smaller than in human epithelial or endothelial cells, due to the lack of Alu elements in mouse genome. Furthermore, during the course of C. albicans infection we observed stable level of A-to-I editing activity in 131 and 190 common editing sites in the mouse tongue and kidney tissues, and found no significant change in ADAR1 or ADAR2 expression (with the exception of ADAR2 displaying a significant increase at 12 h after infection in mouse kidney tissue before returning to normal).
Conclusions: This work represents the first comprehensive analysis of A-to-I RNA editome in human epithelial and endothelial cells. C. albicans infection of human epithelial and endothelial cells led to the up-regulation of A-to-I editing activities, through a mechanism different from that of viral infections in human hosts. However, the in vivo mouse model with C. albicans infection did not show significant changes in A-to-I editing activities in tongue and kidney tissues. The different results in the mouse model were likely due to the presence of more complex in vivo environments, e.g. circulation and mixed cell types.
Keywords: A-to-I RNA-editing; ADAR; Candida albicans; Fungi-host interaction; Infection.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
A comprehensive study on cellular RNA editing activity in response to infections with different subtypes of influenza a viruses.BMC Genomics. 2018 Jan 19;19(Suppl 1):925. doi: 10.1186/s12864-017-4330-1. BMC Genomics. 2018. PMID: 29363430 Free PMC article.
-
Adenosine-to-inosine Alu RNA editing controls the stability of the pro-inflammatory long noncoding RNA NEAT1 in atherosclerotic cardiovascular disease.J Mol Cell Cardiol. 2021 Nov;160:111-120. doi: 10.1016/j.yjmcc.2021.07.005. Epub 2021 Jul 21. J Mol Cell Cardiol. 2021. PMID: 34302813 Free PMC article.
-
Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing.PLoS One. 2010 Jun 21;5(6):e11173. doi: 10.1371/journal.pone.0011173. PLoS One. 2010. PMID: 20574523 Free PMC article.
-
A-to-I RNA editing and hematopoiesis.Exp Hematol. 2024 Nov;139:104621. doi: 10.1016/j.exphem.2024.104621. Epub 2024 Aug 24. Exp Hematol. 2024. PMID: 39187172 Review.
-
In cancer, A-to-I RNA editing can be the driver, the passenger, or the mechanic.Drug Resist Updat. 2017 May;32:16-22. doi: 10.1016/j.drup.2017.09.001. Epub 2017 Oct 4. Drug Resist Updat. 2017. PMID: 29145975 Review.
Cited by
-
Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes.EMBO Rep. 2021 May 5;22(5):e52255. doi: 10.15252/embr.202052255. Epub 2021 Mar 1. EMBO Rep. 2021. PMID: 33645898 Free PMC article.
-
Host A-to-I RNA editing signatures in intracellular bacterial and single-strand RNA viral infections.Front Immunol. 2023 Apr 4;14:1121096. doi: 10.3389/fimmu.2023.1121096. eCollection 2023. Front Immunol. 2023. PMID: 37081881 Free PMC article.
-
A bioinformatics potpourri.BMC Genomics. 2018 Jan 19;19(Suppl 1):920. doi: 10.1186/s12864-017-4326-x. BMC Genomics. 2018. PMID: 29363432 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

