Probing the Mechanism of Receptor Activity-Modifying Protein Modulation of GPCR Ligand Selectivity through Rational Design of Potent Adrenomedullin and Calcitonin Gene-Related Peptide Antagonists
- PMID: 29363552
- PMCID: PMC5832325
- DOI: 10.1124/mol.117.110916
Probing the Mechanism of Receptor Activity-Modifying Protein Modulation of GPCR Ligand Selectivity through Rational Design of Potent Adrenomedullin and Calcitonin Gene-Related Peptide Antagonists
Abstract
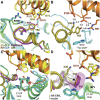
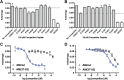
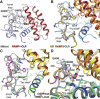
Binding of the vasodilator peptides adrenomedullin (AM) and calcitonin gene-related peptide (CGRP) to the class B G protein-coupled receptor calcitonin receptor-like receptor (CLR) is modulated by receptor activity-modifying proteins (RAMPs). RAMP1 favors CGRP, whereas RAMP2 and RAMP3 favor AM. Crystal structures of peptide-bound RAMP1/2-CLR extracellular domain (ECD) heterodimers suggested RAMPs alter ligand preference through direct peptide contacts and allosteric modulation of CLR. Here, we probed this dual mechanism through rational structure-guided design of AM and CGRP antagonist variants. Variants were characterized for binding to purified RAMP1/2-CLR ECD and for antagonism of the full-length CGRP (RAMP1:CLR), AM1 (RAMP2:CLR), and AM2 (RAMP3:CLR) receptors. Short nanomolar affinity AM(37-52) and CGRP(27-37) variants were obtained through substitutions including AM S45W/Q50W and CGRP K35W/A36S designed to stabilize their β-turn. K46L and Y52F substitutions designed to exploit RAMP allosteric effects and direct peptide contacts, respectively, yielded AM variants with selectivity for the CGRP receptor over the AM1 receptor. AM(37-52) S45W/K46L/Q50W/Y52F exhibited nanomolar potency at the CGRP receptor and micromolar potency at AM1 A 2.8-Å resolution crystal structure of this variant bound to the RAMP1-CLR ECD confirmed that it bound as designed. CGRP(27-37) N31D/S34P/K35W/A36S exhibited potency and selectivity comparable to the traditional antagonist CGRP(8-37). Giving this variant the ability to contact RAMP2 through the F37Y substitution increased affinity for AM1, but it still preferred the CGRP receptor. These potent peptide antagonists with altered selectivity inform the development of AM/CGRP-based pharmacological tools and support the hypothesis that RAMPs alter CLR ligand selectivity through allosteric effects and direct peptide contacts.
Copyright © 2018 by The American Society for Pharmacology and Experimental Therapeutics.
Figures






Similar articles
-
Structure-function analyses reveal a triple β-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design.J Biol Chem. 2018 Oct 12;293(41):15840-15854. doi: 10.1074/jbc.RA118.005062. Epub 2018 Aug 23. J Biol Chem. 2018. PMID: 30139742 Free PMC article.
-
Receptor activity-modifying protein-dependent effects of mutations in the calcitonin receptor-like receptor: implications for adrenomedullin and calcitonin gene-related peptide pharmacology.Br J Pharmacol. 2014 Feb;171(3):772-88. doi: 10.1111/bph.12508. Br J Pharmacol. 2014. PMID: 24199627 Free PMC article.
-
Cardiovascular effects of exogenous adrenomedullin and CGRP in Ramp and Calcrl deficient mice.Peptides. 2017 Feb;88:1-7. doi: 10.1016/j.peptides.2016.12.002. Epub 2016 Dec 8. Peptides. 2017. PMID: 27940069 Free PMC article.
-
Shared and separate functions of the RAMP-based adrenomedullin receptors.Peptides. 2011 Jul;32(7):1540-50. doi: 10.1016/j.peptides.2011.05.022. Epub 2011 May 27. Peptides. 2011. PMID: 21645567 Review.
-
Ectodomain structures of the CGRP and AM receptors.Curr Protein Pept Sci. 2013 Aug;14(5):375-85. doi: 10.2174/13892037113149990054. Curr Protein Pept Sci. 2013. PMID: 23745701 Review.
Cited by
-
Molecular interaction of an antagonistic amylin analog with the extracellular domain of receptor activity-modifying protein 2 assessed by fluorescence polarization.Biophys Chem. 2020 Dec;267:106477. doi: 10.1016/j.bpc.2020.106477. Epub 2020 Sep 20. Biophys Chem. 2020. PMID: 33137565 Free PMC article.
-
Asn-linked N-acetylglucosamine of the amylin receptor 2 extracellular domain enhances peptide ligand affinity.FEBS Open Bio. 2021 Jan;11(1):195-206. doi: 10.1002/2211-5463.13042. Epub 2020 Dec 2. FEBS Open Bio. 2021. PMID: 33227824 Free PMC article.
-
Calcitonin Receptor N-Glycosylation Enhances Peptide Hormone Affinity by Controlling Receptor Dynamics.J Mol Biol. 2020 Mar 27;432(7):1996-2014. doi: 10.1016/j.jmb.2020.01.028. Epub 2020 Feb 6. J Mol Biol. 2020. PMID: 32035902 Free PMC article.
-
Development of High Affinity Calcitonin Analog Fragments Targeting Extracellular Domains of Calcitonin Family Receptors.Biomolecules. 2021 Sep 15;11(9):1364. doi: 10.3390/biom11091364. Biomolecules. 2021. PMID: 34572577 Free PMC article.
-
Pipeline for development of acylated peptide based CGRP receptor antagonist with extended half-life for migraine treatment.Sci Rep. 2025 Jan 13;15(1):1870. doi: 10.1038/s41598-024-84547-1. Sci Rep. 2025. PMID: 39805895 Free PMC article.
References
-
- Aubdool AA, Thakore P, Argunhan F, Smillie SJ, Schnelle M, Srivastava S, Alawi KM, Wilde E, Mitchell J, Farrell-Dillon K, et al. (2017) A novel α-calcitonin gene-related peptide analogue protects against end-organ damage in experimental hypertension, cardiac hypertrophy, and heart failure. Circulation 136:367–383. - PMC - PubMed
-
- Bailey RJ, Hay DL. (2006) Pharmacology of the human CGRP1 receptor in Cos 7 cells. Peptides 27:1367–1375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

