Structural basis of the specific interactions of GRAS family proteins
- PMID: 29364510
- PMCID: PMC5873383
- DOI: 10.1002/1873-3468.12987
Structural basis of the specific interactions of GRAS family proteins
Abstract
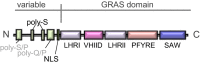
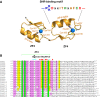
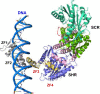
The plant-specific GAI-RGA-and-SCR (GRAS) family of proteins function as transcriptional regulators and play critical roles in development and signalling. Recent structural studies have shed light on the molecular functions at the structural level. The conserved GRAS domain comprises an α-helical cap and α/β core subdomains. The α-helical cap mediates head-to-head heterodimerization between SHR and SCR GRAS domains. This type of dimerization is predicted for the NSP1-NSP2 heterodimer and DELLA proteins such as RGA and SLR1 homodimers. The α/β core subdomain possesses a hydrophobic groove formed by surface α3- and α7-helices and mediates protein-protein interactions. The groove of the SHR GRAS domain accommodates the zinc fingers of JKD, a BIRD/IDD family transcription factor, while the groove of the SCL7 GRAS domain mediates the SCL7 homodimerization.
Keywords: BIRD family; DELLA protein; GRAS domain; IDD family; SAM-dependent methyltransferase; Transcription cofactor; Transcription factor; Zinc finger; α/β protein.
© 2018 The Author. FEBS Letters published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Figures






Similar articles
-
Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD.Nat Plants. 2017 Feb 17;3:17010. doi: 10.1038/nplants.2017.10. Nat Plants. 2017. PMID: 28211915 Free PMC article.
-
Crystal Structure of the GRAS Domain of SCARECROW-LIKE7 in Oryza sativa.Plant Cell. 2016 May;28(5):1025-34. doi: 10.1105/tpc.16.00018. Epub 2016 Apr 14. Plant Cell. 2016. PMID: 27081181 Free PMC article.
-
Expression and purification of a GRAS domain of SLR1, the rice DELLA protein.Protein Expr Purif. 2014 Mar;95:248-58. doi: 10.1016/j.pep.2014.01.006. Epub 2014 Jan 23. Protein Expr Purif. 2014. PMID: 24463428
-
GRAS-domain transcription factors that regulate plant development.Plant Signal Behav. 2009 Aug;4(8):698-700. doi: 10.4161/psb.4.8.9176. Epub 2009 Aug 4. Plant Signal Behav. 2009. PMID: 19820314 Free PMC article. Review.
-
The role of GRAS proteins in plant signal transduction and development.Planta. 2004 Mar;218(5):683-92. doi: 10.1007/s00425-004-1203-z. Epub 2004 Feb 4. Planta. 2004. PMID: 14760535 Review.
Cited by
-
Transcriptomic analysis reveals the GRAS family genes respond to gibberellin in Salvia miltiorrhiza hairy roots.BMC Genomics. 2020 Oct 27;21(1):727. doi: 10.1186/s12864-020-07119-3. BMC Genomics. 2020. PMID: 33106159 Free PMC article.
-
Early "Rootprints" of Plant Terrestrialization: Selaginella Root Development Sheds Light on Root Evolution in Vascular Plants.Front Plant Sci. 2021 Sep 4;12:735514. doi: 10.3389/fpls.2021.735514. eCollection 2021. Front Plant Sci. 2021. PMID: 34671375 Free PMC article.
-
Cellular and physiological functions of SGR family in gravitropic response in higher plants.J Adv Res. 2025 Jan;67:43-60. doi: 10.1016/j.jare.2024.01.026. Epub 2024 Feb 1. J Adv Res. 2025. PMID: 38295878 Free PMC article. Review.
-
Genome-Wide Identification and Expression Pattern of the GRAS Gene Family in Pitaya (Selenicereus undatus L.).Biology (Basel). 2022 Dec 21;12(1):11. doi: 10.3390/biology12010011. Biology (Basel). 2022. PMID: 36671704 Free PMC article.
-
SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice.Nat Commun. 2019 Jun 21;10(1):2738. doi: 10.1038/s41467-019-10667-2. Nat Commun. 2019. PMID: 31227696 Free PMC article.
References
-
- Pysh LD, Wysocka‐Diller JW, Camilleri C, Bouchez D and Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW‐LIKE genes. Plant J 18, 111–119. - PubMed
-
- Sun TP (2011) The molecular mechanism and evolution of the GA‐GID1‐DELLA signaling module in plants. Curr Biol 21, R338–R345. - PubMed
-
- Pauluzzi G, Divol F, Puig J, Guiderdoni E, Dievart A & Périn C (2012) Surfing along the root ground tissue gene network. Dev Biol 365, 14–22. - PubMed
-
- Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218, 683–692. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

