OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice
- PMID: 29364524
- PMCID: PMC5873253
- DOI: 10.1111/nph.14977
OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice
Abstract
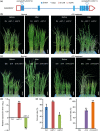
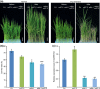
Plants modify their development to adapt to their environment, protecting themselves from detrimental conditions such as chilling stress by triggering a variety of signaling pathways; however, little is known about how plants coordinate developmental patterns and stress responses at the molecular level. Here, we demonstrate that interacting transcription factors OsMADS57 and OsTB1 directly target the defense gene OsWRKY94 and the organogenesis gene D14 to trade off the functions controlling/moderating rice tolerance to cold. Overexpression of OsMADS57 maintains rice tiller growth under chilling stress. OsMADS57 binds directly to the promoter of OsWRKY94, activating its transcription for the cold stress response, while suppressing its activity under normal temperatures. In addition, OsWRKY94 was directly targeted and suppressed by OsTB1 under both normal and chilling temperatures. However, D14 transcription was directly promoted by OsMADS57 for suppressing tillering under the chilling treatment, whereas D14 was repressed for enhancing tillering under normal condition.We demonstrated that OsMADS57 and OsTB1 conversely affect rice chilling tolerance via targeting OsWRKY94. Our findings highlight a molecular genetic mechanism coordinating organogenesis and chilling tolerance in rice, which supports and extends recent work suggesting that chilling stress environments influence organ differentiation.
Keywords: D14; OsMADS57; WRKY94; cold tolerance; gene network; organogenesis; rice (Oryza sativa); trade-off.
© 2018 The Authors. New Phytologist © 2018 New Phytologist Trust.
Figures



Similar articles
-
The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14.Nat Commun. 2013;4:1566. doi: 10.1038/ncomms2542. Nat Commun. 2013. PMID: 23463009 Free PMC article.
-
Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice.Mol Plant. 2021 Feb 1;14(2):315-329. doi: 10.1016/j.molp.2020.11.022. Epub 2020 Dec 2. Mol Plant. 2021. PMID: 33278597
-
A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants.Physiol Plant. 2021 Nov;173(3):1120-1135. doi: 10.1111/ppl.13508. Epub 2021 Jul 29. Physiol Plant. 2021. PMID: 34287928
-
Coordination of light, circadian clock with temperature: The potential mechanisms regulating chilling tolerance in rice.J Integr Plant Biol. 2020 Jun;62(6):737-760. doi: 10.1111/jipb.12852. Epub 2019 Sep 18. J Integr Plant Biol. 2020. PMID: 31243851 Review.
-
Chilling tolerance in rice: Past and present.J Plant Physiol. 2022 Jan;268:153576. doi: 10.1016/j.jplph.2021.153576. Epub 2021 Dec 1. J Plant Physiol. 2022. PMID: 34875419 Review.
Cited by
-
The Role of Dwarfing Traits in Historical and Modern Agriculture with a Focus on Rice.Cold Spring Harb Perspect Biol. 2019 Nov 1;11(11):a034645. doi: 10.1101/cshperspect.a034645. Cold Spring Harb Perspect Biol. 2019. PMID: 31358515 Free PMC article. Review.
-
Alleviation of drought stress in tomato by foliar application of seafood waste extract.Sci Rep. 2024 Dec 20;14(1):30572. doi: 10.1038/s41598-024-80798-0. Sci Rep. 2024. PMID: 39706919 Free PMC article.
-
Mutagenesis in Rice: The Basis for Breeding a New Super Plant.Front Plant Sci. 2019 Nov 8;10:1326. doi: 10.3389/fpls.2019.01326. eCollection 2019. Front Plant Sci. 2019. PMID: 31781133 Free PMC article. Review.
-
Rice functional genomics: decades' efforts and roads ahead.Sci China Life Sci. 2022 Jan;65(1):33-92. doi: 10.1007/s11427-021-2024-0. Epub 2021 Dec 7. Sci China Life Sci. 2022. PMID: 34881420 Review.
-
Genome-wide identification of the plant homeodomain-finger family in rye and ScPHD5 functions in cold tolerance and flowering time.Plant Cell Rep. 2024 May 15;43(6):142. doi: 10.1007/s00299-024-03226-7. Plant Cell Rep. 2024. PMID: 38744747
References
-
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. 2007. DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant Journal 51: 1019–1029. - PubMed
-
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J. 2009. d14, a strigolactone‐insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiology 50: 1416–1424. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

