Genetic landscape of ultra-stable chronic lymphocytic leukemia patients
- PMID: 29365086
- PMCID: PMC6248613
- DOI: 10.1093/annonc/mdy021
Genetic landscape of ultra-stable chronic lymphocytic leukemia patients
Abstract
Background: Chronic lymphocytic leukemia (CLL) has a heterogeneous clinical course. Beside patients requiring immediate treatment, others show an initial indolent phase followed by progression and others do not progress for decades. The latter two subgroups usually display mutated IGHV genes and a favorable FISH profile.
Patients and methods: Patients with absence of disease progression for over 10 years (10-34) from diagnosis were defined as ultra-stable CLL (US-CLL). Forty US-CLL underwent extensive characterization including whole exome sequencing (WES), ultra-deep sequencing and copy number aberration (CNA) analysis to define their unexplored genetic landscape. Microarray analysis, comparing US-CLL with non-US-CLL with similar immunogenetic features (mutated IGHV/favorable FISH), was also carried out to recognize US-CLL at diagnosis.
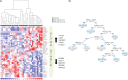
Results: WES was carried out in 20 US-CLL and 84 non-silent somatic mutations in 78 genes were found. When re-tested in a validation cohort of 20 further US-CLL, no recurrent lesion was identified. No clonal mutations of NOTCH1, BIRC3, SF3B1 and TP53 were found, including ATM and other potential progression driving mutations. CNA analysis identified 31 lesions, none with known poor prognostic impact. No novel recurrent lesion was identified: most cases showed no lesions (38%) or an isolated del(13q) (31%). The expression of 6 genes, selected from a gene expression profile analysis by microarray and quantified by droplet digital PCR on a cohort of 79 CLL (58 US-CLL and 21 non-US-CLL), allowed to build a decision-tree capable of recognizing at diagnosis US-CLL patients.
Conclusions: The genetic landscape of US-CLL is characterized by the absence of known unfavorable driver mutations/CNA and of novel recurrent genetic lesions. Among CLL patients with favorable immunogenetics, a decision-tree based on the expression of 6 genes may identify at diagnosis patients who are likely to maintain an indolent disease for decades.
Figures

References
-
- Fabbri G, Dalla-Favera R.. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016; 16(3): 145–162 (Review). - PubMed
-
- Pospisilova S, Gonzalez D, Malcikova J. et al. ; European Research Initiative on CLL (ERIC). ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia 2012; 26(7): 1458–1461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

