Non-B-Form DNA Is Enriched at Centromeres
- PMID: 29365169
- PMCID: PMC5889037
- DOI: 10.1093/molbev/msy010
Non-B-Form DNA Is Enriched at Centromeres
Erratum in
-
Molecular Biology and Evolution, Volume 35, Issue 4.Mol Biol Evol. 2018 Jul 1;35(7):1821. doi: 10.1093/molbev/msy057. Mol Biol Evol. 2018. PMID: 29659984 Free PMC article. No abstract available.
Abstract
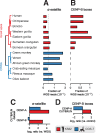
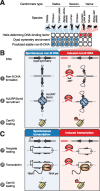
Animal and plant centromeres are embedded in repetitive "satellite" DNA, but are thought to be epigenetically specified. To define genetic characteristics of centromeres, we surveyed satellite DNA from diverse eukaryotes and identified variation in <10-bp dyad symmetries predicted to adopt non-B-form conformations. Organisms lacking centromeric dyad symmetries had binding sites for sequence-specific DNA-binding proteins with DNA-bending activity. For example, human and mouse centromeres are depleted for dyad symmetries, but are enriched for non-B-form DNA and are associated with binding sites for the conserved DNA-binding protein CENP-B, which is required for artificial centromere function but is paradoxically nonessential. We also detected dyad symmetries and predicted non-B-form DNA structures at neocentromeres, which form at ectopic loci. We propose that centromeres form at non-B-form DNA because of dyad symmetries or are strengthened by sequence-specific DNA binding proteins. This may resolve the CENP-B paradox and provide a general basis for centromere specification.
Figures




References
-
- Arnold C, Matthews LJ, Nunn CL.. 2010. The 10kTrees website: a new online resource for primate phylogeny. Evol Anthropol. 193:114–118. 10.1002/evan.20251 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

