Epstein-Barr Virus (EBV) Latent Protein EBNA3A Directly Targets and Silences the STK39 Gene in B Cells Infected by EBV
- PMID: 29367247
- PMCID: PMC5972881
- DOI: 10.1128/JVI.01918-17
Epstein-Barr Virus (EBV) Latent Protein EBNA3A Directly Targets and Silences the STK39 Gene in B Cells Infected by EBV
Abstract
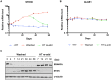
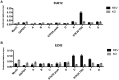
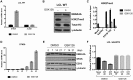
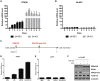
Epstein-Barr virus (EBV) establishes latent infection in human B cells and is associated with a wide range of cancers. The EBV nuclear antigen 3 (EBNA3) family proteins are critical for B cell transformation and function as transcriptional regulators. It is well established that EBNA3A and EBNA3C cooperate in the regulation of cellular genes. Here, we demonstrate that the gene STK39 is repressed only by EBNA3A. This is the first example of a gene regulated only by EBNA3A in EBV-transformed lymphoblastoid cell lines (LCLs) without the help of EBNA3C. This was demonstrated using a variety of LCLs carrying either knockout, revertant, or conditional EBNA3 recombinants. Investigating the kinetics of EBNA3A-mediated changes in STK39 expression showed that STK39 becomes derepressed quickly after EBNA3A inactivation. This derepression is reversible as EBNA3A reactivation represses STK39 in the same cells expressing a conditional EBNA3A. STK39 is silenced shortly after primary B cell infection by EBV, and no STK39-encoded protein (SPAK) is detected 3 weeks postinfection. Chromatin immunoprecipitation (ChIP) analysis indicates that EBNA3A directly binds to a regulatory region downstream of the STK39 transcription start site. For the first time, we demonstrated that the polycomb repressive complex 2 with the deposition of the repressive mark H3K27me3 is not only important for the maintenance of an EBNA3A target gene (STK39) but is also essential for the initial establishment of its silencing. Finally, we showed that DNA methyltransferases are involved in the EBNA3A-mediated repression of STK39IMPORTANCE EBV is well known for its ability to transform B lymphocytes to continuously proliferating lymphoblastoid cell lines. This is achieved in part by the reprogramming of cellular gene transcription by EBV transcription factors, including the EBNA3 proteins that play a crucial role in this process. In the present study, we found that EBNA3A epigenetically silences STK39 This is the first gene where EBNA3A has been found to exert its repressive role by itself, without needing its coregulators EBNA3B and EBNA3C. Furthermore, we demonstrated that the polycomb repressor complex is essential for EBNA3A-mediated repression of STK39 Findings in this study provide new insights into the regulation of cellular genes by the transcription factor EBNA3A.
Keywords: EBNA3; Epstein-Barr virus; transcriptional regulation; virology.
Copyright © 2018 Bazot et al.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

