Mimicry of Cellular A Kinase-Anchoring Proteins Is a Conserved and Critical Function of E1A across Various Human Adenovirus Species
- PMID: 29367252
- PMCID: PMC5874402
- DOI: 10.1128/JVI.01902-17
Mimicry of Cellular A Kinase-Anchoring Proteins Is a Conserved and Critical Function of E1A across Various Human Adenovirus Species
Abstract
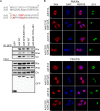
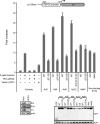
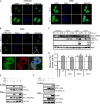
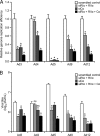
The E1A proteins of the various human adenovirus (HAdV) species perform the critical task of converting an infected cell into a setting primed for virus replication. While E1A proteins differ in both sequence and mechanism, the evolutionary pressure on viruses with limited coding capacity ensures that these proteins often have significant overlap in critical functions. HAdV-5 E1A is known to use mimicry to rewire cyclic AMP (cAMP) signaling by decoupling protein kinase A (PKA) from cellular A kinase-anchoring proteins (AKAPs) and utilizing PKA to its own advantage. We show here that E1As from other species of HAdV also possess this viral AKAP (vAKAP) function and examine how they manipulate PKA. E1A from most species of HAdV examined contain a small AKAP-like motif in their N terminus which targets the docking-dimerization domain of PKA as the binding interface for a conserved protein-protein interaction. This motif is also responsible for an E1A-mediated relocalization of PKA regulatory subunits from the cytoplasm into the nucleus, with species-specific E1A proteins having preference for one particular isoform of PKA subunit over another. Importantly, we showed that these newly characterized vAKAPs can integrate into cAMP-responsive transcription as well as contribute to viral genome replication and infectious progeny production for several distinct HAdV species.IMPORTANCE These data enhance the mechanistic knowledge on how HAdV E1A manipulates cellular PKA to benefit infection. The work establishes that mimicry of AKAPs and subversion of PKA-mediated cAMP signaling are conserved features for numerous human adenoviruses. This study also highlights the molecular determinants conferring selective protein-protein interactions between distinct PKA regulatory subunits and the different E1A proteins of these viruses. Additionally, it further emphasizes the utility of using viral proteins like E1A as tools for studying the molecular biology of cellular regulatory pathways.
Keywords: A-kinase anchoring protein; E1A; adenovirus; cyclic AMP; evolution; protein kinase A; protein-protein interaction; replication; viral AKAP; viral mimicry.
Copyright © 2018 American Society for Microbiology.
Figures



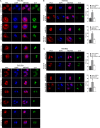
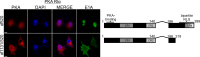


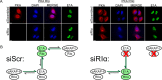
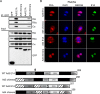
References
-
- Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, Berk AJ. 2014. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16:663–676. doi:10.1016/j.chom.2014.10.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

