Interregulation of CDKA/CDK1 and the Plant-Specific Cyclin-Dependent Kinase CDKB in Control of the Chlamydomonas Cell Cycle
- PMID: 29367304
- PMCID: PMC5868683
- DOI: 10.1105/tpc.17.00759
Interregulation of CDKA/CDK1 and the Plant-Specific Cyclin-Dependent Kinase CDKB in Control of the Chlamydomonas Cell Cycle
Abstract
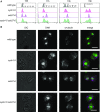
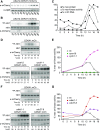
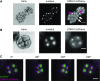
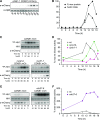
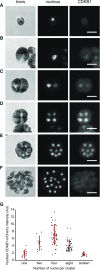
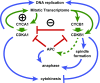
The cyclin-dependent kinase CDK1 is essential for mitosis in fungi and animals. Plant genomes contain the CDK1 ortholog CDKA and a plant kingdom-specific relative, CDKB. The green alga Chlamydomonas reinhardtii has a long G1 growth period followed by rapid cycles of DNA replication and cell division. We show that null alleles of CDKA extend the growth period prior to the first division cycle and modestly extend the subsequent division cycles, but do not prevent cell division, indicating at most a minor role for the CDK1 ortholog in mitosis in Chlamydomonas. A null allele of cyclin A has a similar though less extreme phenotype. In contrast, both CDKB and cyclin B are essential for mitosis. CDK kinase activity measurements imply that the predominant in vivo complexes are probably cyclin A-CDKA and cyclin B-CDKB. We propose a negative feedback loop: CDKA activates cyclin B-CDKB. Cyclin B-CDKB in turn promotes mitotic entry and inactivates cyclin A-CDKA. Cyclin A-CDKA and cyclin B-CDKB may redundantly promote DNA replication. We show that the anaphase-promoting complex is required for inactivation of both CDKA and CDKB and is essential for anaphase. These results are consistent with findings in Arabidopsis thaliana and may delineate the core of plant kingdom cell cycle control that, compared with the well-studied yeast and animal systems, exhibits deep conservation in some respects and striking divergence in others.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures



Comment in
-
Cell Cycle Regulation by Chlamydomonas Cyclin-Dependent Protein Kinases.Plant Cell. 2018 Feb;30(2):271. doi: 10.1105/tpc.18.00103. Epub 2018 Feb 5. Plant Cell. 2018. PMID: 29437987 Free PMC article. No abstract available.
References
-
- Bloom J., Cross F.R. (2007). Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 8: 149–160. - PubMed
-
- Boudolf V., Vlieghe K., Beemster G.T.S., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

