Role of the Nod Factor Hydrolase MtNFH1 in Regulating Nod Factor Levels during Rhizobial Infection and in Mature Nodules of Medicago truncatula
- PMID: 29367305
- PMCID: PMC5868697
- DOI: 10.1105/tpc.17.00420
Role of the Nod Factor Hydrolase MtNFH1 in Regulating Nod Factor Levels during Rhizobial Infection and in Mature Nodules of Medicago truncatula
Abstract
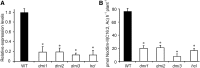
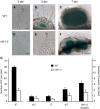
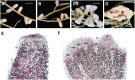
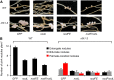
Establishment of symbiosis between legumes and nitrogen-fixing rhizobia depends on bacterial Nod factors (NFs) that trigger symbiosis-related NF signaling in host plants. NFs are modified oligosaccharides of chitin with a fatty acid moiety. NFs can be cleaved and inactivated by host enzymes, such as MtNFH1 (MEDICAGO TRUNCATULA NOD FACTOR HYDROLASE1). In contrast to related chitinases, MtNFH1 hydrolyzes neither chitin nor chitin fragments, indicating a high cleavage preference for NFs. Here, we provide evidence for a role of MtNFH1 in the symbiosis with Sinorhizobium meliloti Upon rhizobial inoculation, MtNFH1 accumulated at the curled tip of root hairs, in the so-called infection chamber. Mutant analysis revealed that lack of MtNFH1 delayed rhizobial root hair infection, suggesting that excess amounts of NFs negatively affect the initiation of infection threads. MtNFH1 deficiency resulted in nodule hypertrophy and abnormal nodule branching of young nodules. Nodule branching was also stimulated in plants expressing MtNFH1 driven by a tandem CaMV 35S promoter and plants inoculated by a NF-overproducing S. meliloti strain. We suggest that fine-tuning of NF levels by MtNFH1 is necessary for optimal root hair infection as well as for NF-regulated growth of mature nodules.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures





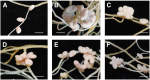
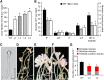
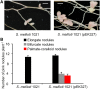
Comment in
-
Goldilocks Principle: MtNFH1 Ensures Optimal Nod Factor Activity.Plant Cell. 2018 Feb;30(2):267-268. doi: 10.1105/tpc.18.00114. Epub 2018 Feb 7. Plant Cell. 2018. PMID: 29436475 Free PMC article. No abstract available.
References
-
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367. - PubMed
-
- Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374. - PMC - PubMed
-
- Auriac M.C., Timmers A.C.J. (2007). Nodulation studies in the model legume Medicago truncatula: advantages of using the constitutive EF1α promoter and limitations in detecting fluorescent reporter proteins in nodule tissues. Mol. Plant Microbe Interact. 20: 1040–1047. - PubMed
-
- Baev N., Endre G., Petrovics G., Banfalvi Z., Kondorosi A. (1991). Six nodulation genes of nod box locus 4 in Rhizobium meliloti are involved in nodulation signal production: nodM codes for D-glucosamine synthetase. Mol. Gen. Genet. 228: 113–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

