Systems analysis of the glycoside hydrolase family 18 enzymes from Cellvibrio japonicus characterizes essential chitin degradation functions
- PMID: 29367339
- PMCID: PMC5846145
- DOI: 10.1074/jbc.RA117.000849
Systems analysis of the glycoside hydrolase family 18 enzymes from Cellvibrio japonicus characterizes essential chitin degradation functions
Abstract
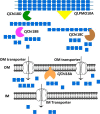
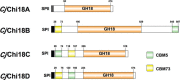
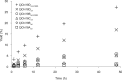
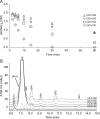
Understanding the strategies used by bacteria to degrade polysaccharides constitutes an invaluable tool for biotechnological applications. Bacteria are major mediators of polysaccharide degradation in nature; however, the complex mechanisms used to detect, degrade, and consume these substrates are not well-understood, especially for recalcitrant polysaccharides such as chitin. It has been previously shown that the model bacterial saprophyte Cellvibrio japonicus is able to catabolize chitin, but little is known about the enzymatic machinery underlying this capability. Previous analyses of the C. japonicus genome and proteome indicated the presence of four glycoside hydrolase family 18 (GH18) enzymes, and studies of the proteome indicated that all are involved in chitin utilization. Using a combination of in vitro and in vivo approaches, we have studied the roles of these four chitinases in chitin bioconversion. Genetic analyses showed that only the chi18D gene product is essential for the degradation of chitin substrates. Biochemical characterization of the four enzymes showed functional differences and synergistic effects during chitin degradation, indicating non-redundant roles in the cell. Transcriptomic studies revealed complex regulation of the chitin degradation machinery of C. japonicus and confirmed the importance of CjChi18D and CjLPMO10A, a previously characterized chitin-active enzyme. With this systems biology approach, we deciphered the physiological relevance of the glycoside hydrolase family 18 enzymes for chitin degradation in C. japonicus, and the combination of in vitro and in vivo approaches provided a comprehensive understanding of the initial stages of chitin degradation by this bacterium.
Keywords: Cellvibrio japonicus; bacterial genetics; bacterial metabolism; chitin; chitinase; glycoside hydrolase; polysaccharide.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Gooday G. W., Humphreys A. M., and McIntosh W. H. (1986) Roles of chitinases in fungal growth. In Chitin in Nature and Technology (Muzzarelli R., Jeuniaux C., and Gooday G. W., eds.) pp. 83–91, Springer-Verlag, New York Inc., New York
-
- Francesko A., and Tzanov T. (2011) Chitin, chitosan and derivatives for wound healing and tissue engineering. Adv. Biochem. Eng. Biotechnol. 125, 1–27 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

