Defining the binding determinants of Shewanella oneidensis OxyR: Implications for the link between the contracted OxyR regulon and adaptation
- PMID: 29367341
- PMCID: PMC5858001
- DOI: 10.1074/jbc.RA117.001530
Defining the binding determinants of Shewanella oneidensis OxyR: Implications for the link between the contracted OxyR regulon and adaptation
Abstract
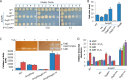
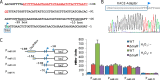
It is well-established that OxyR functions as a transcriptional activator of the peroxide stress response in bacteria, primarily based on studies on Escherichia coli Recent investigations have revealed that OxyRs of some other bacteria can regulate gene expression through both repression and activation or repression only; however, the underlying mechanisms remain largely unknown. Here, we demonstrated in γ-proteobacteriumShewanella oneidensis regulation of OxyR on expression of major catalase gene katB in a dual-control manner through interaction with a single site in the promoter region. Under non-stress conditions, katB expression was repressed by reduced OxyR (OxyRred), whereas when oxidized, OxyR (OxyRoxi) outcompeted OxyRred for the site because of substantially enhanced affinity, resulting in a graded response to oxidative stress, from repression to derepression to activation. The OxyR-binding motif is characterized as a combination of the E. coli motif (tetranucleotides spaced by heptanucleotide) and palindromic structure. We provided evidence to suggest that the S. oneidensis OxyR regulon is significantly contracted compared with those reported, probably containing only five members that are exclusively involved in oxygen reactive species scavenging and iron sequestering. These characteristics probably reflect the adapting strategy of the bacteria that S. oneidensis represents to thrive in redox-stratified microaerobic and anaerobic environments.
Keywords: DNA binding protein; bacterial genetics; gene regulation; hydrogen peroxide; oxidative stress.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Managing oxidative stresses in Shewanella oneidensis: intertwined roles of the OxyR and OhrR regulons.Environ Microbiol. 2014 Jun;16(6):1821-34. doi: 10.1111/1462-2920.12418. Environ Microbiol. 2014. PMID: 25009841
-
The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis.J Bacteriol. 2000 Sep;182(18):5059-69. doi: 10.1128/JB.182.18.5059-5069.2000. J Bacteriol. 2000. PMID: 10960088 Free PMC article.
-
Unraveling the Mechanism for the Viability Deficiency of Shewanella oneidensis oxyR Null Mutant.J Bacteriol. 2015 Jul;197(13):2179-2189. doi: 10.1128/JB.00154-15. Epub 2015 Apr 20. J Bacteriol. 2015. PMID: 25897035 Free PMC article.
-
The OxyR regulon.Antonie Van Leeuwenhoek. 1990 Oct;58(3):157-61. doi: 10.1007/BF00548927. Antonie Van Leeuwenhoek. 1990. PMID: 2256675 Review.
-
Regulation of hydroperoxidase (catalase) expression in Escherichia coli.FEMS Microbiol Lett. 1995 Sep 1;131(2):113-9. doi: 10.1111/j.1574-6968.1995.tb07764.x. FEMS Microbiol Lett. 1995. PMID: 7557318 Review.
Cited by
-
Microbial helpers allow cyanobacteria to thrive in ferruginous waters.Geobiology. 2021 Sep;19(5):510-520. doi: 10.1111/gbi.12443. Epub 2021 Apr 19. Geobiology. 2021. PMID: 33871172 Free PMC article.
-
Functional Irreplaceability of Escherichia coli and Shewanella oneidensis OxyRs Is Critically Determined by Intrinsic Differences in Oligomerization.mBio. 2022 Feb 22;13(1):e0349721. doi: 10.1128/mbio.03497-21. Epub 2022 Jan 25. mBio. 2022. PMID: 35073744 Free PMC article.
-
Structural and functional insights into transcription activation of the essential LysR-type transcriptional regulators.Protein Sci. 2024 Jun;33(6):e5012. doi: 10.1002/pro.5012. Protein Sci. 2024. PMID: 38723180 Free PMC article.
-
Bacterial TANGO2 homologs are heme-trafficking proteins that facilitate biosynthesis of cytochromes c.mBio. 2023 Aug 31;14(4):e0132023. doi: 10.1128/mbio.01320-23. Epub 2023 Jul 18. mBio. 2023. PMID: 37462360 Free PMC article.
-
Distinct H2O2-Scavenging System in Yersinia pseudotuberculosis: KatG and AhpC Act Together to Scavenge Endogenous Hydrogen Peroxide.Front Microbiol. 2021 May 7;12:626874. doi: 10.3389/fmicb.2021.626874. eCollection 2021. Front Microbiol. 2021. PMID: 34025596 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

