Loss of eIF4E Phosphorylation Engenders Depression-like Behaviors via Selective mRNA Translation
- PMID: 29367404
- PMCID: PMC5824745
- DOI: 10.1523/JNEUROSCI.2673-17.2018
Loss of eIF4E Phosphorylation Engenders Depression-like Behaviors via Selective mRNA Translation
Abstract
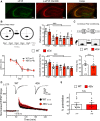
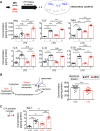
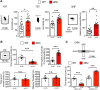
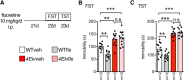
The MAPK/ERK (mitogen-activated protein kinases/extracellular signal-regulated kinase) pathway is a cardinal regulator of synaptic plasticity, learning, and memory in the hippocampus. One of major endpoints of this signaling cascade is the 5' mRNA cap binding protein eIF4E (eukaryotic Initiation Factor 4E), which is phosphorylated on Ser 209 by MNK (MAPK-interacting protein kinases) and controls mRNA translation. The precise role of phospho-eIF4E in the brain is yet to be determined. Herein, we demonstrate that ablation of eIF4E phosphorylation in male mice (4Eki mice) does not impair long-term spatial or contextual fear memory, or the late phase of LTP. Using unbiased translational profiling in mouse brain, we show that phospho-eIF4E differentially regulates the translation of a subset of mRNAs linked to inflammation, the extracellular matrix, pituitary hormones, and the serotonin pathway. Consequently, 4Eki male mice display exaggerated inflammatory responses and reduced levels of serotonin, concomitant with depression and anxiety-like behaviors. Remarkably, eIF4E phosphorylation is required for the chronic antidepressant action of the selective serotonin reuptake inhibitor fluoxetine. Finally, we propose a novel phospho-eIF4E-dependent translational control mechanism in the brain, via the GAIT complex (gamma IFN activated inhibitor of translation). In summary, our work proposes a novel translational control mechanism involved in the regulation of inflammation and depression, which could be exploited to design novel therapeutics.SIGNIFICANCE STATEMENT We demonstrate that downstream of the MAPK (mitogen-activated protein kinase) pathway, eukaryotic Initiation Factor 4E (eIF4E) Ser209 phosphorylation is not required for classical forms of hippocampal LTP and memory. We reveal a novel role for eIF4E phosphorylation in inflammatory responses and depression-like behaviors. eIF4E phosphorylation is required for the chronic action of antidepressants, such as fluoxetine in mice. These phenotypes are accompanied by selective translation of extracellular matrix, pituitary hormones, and serotonin pathway genes, in eIF4E phospho-mutant mice. We also describe a previously unidentified translational control mechanism in the brain, whereby eIF4E phosphorylation is required for inhibiting the translation of gamma IFN activated inhibitor of translation element-containing mRNAs. These findings can be used to design novel therapeutics for depression.
Keywords: depression; eIF4E; inflammation; phospho-eIF4E; translation.
Copyright © 2018 Amorim et al.
Figures


References
-
- Bob P, Fedor-Freybergh PG, Susta M, Pavlat J, Jasova D, Zima T, Benakova H, Miklosko J, Hynek K, Raboch J (2007) Depression, prolactin and dissociated mind. Neuroendocrinol Lett 28:639–642. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
