Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry
- PMID: 29367422
- PMCID: PMC5819400
- DOI: 10.1073/pnas.1714085115
Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry
Abstract
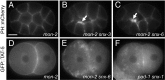
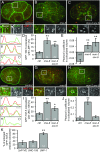
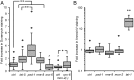
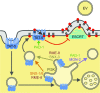
Cells release extracellular vesicles (EVs) that mediate intercellular communication and repair damaged membranes. Despite the pleiotropic functions of EVs in vitro, their in vivo function is debated, largely because it is unclear how to induce or inhibit their formation. In particular, the mechanisms of EV release by plasma membrane budding or ectocytosis are poorly understood. We previously showed that TAT-5 phospholipid flippase activity maintains the asymmetric localization of the lipid phosphatidylethanolamine (PE) in the plasma membrane and inhibits EV budding by ectocytosis in Caenorhabditis elegans However, no proteins that inhibit ectocytosis upstream of TAT-5 were known. Here, we identify TAT-5 regulators associated with retrograde endosomal recycling: PI3Kinase VPS-34, Beclin1 homolog BEC-1, DnaJ protein RME-8, and the uncharacterized Dopey homolog PAD-1. PI3Kinase, RME-8, and semiredundant sorting nexins are required for the plasma membrane localization of TAT-5, which is important to maintain PE asymmetry and inhibit EV release. PAD-1 does not directly regulate TAT-5 localization, but is required for the lipid flipping activity of TAT-5. PAD-1 also has roles in endosomal trafficking with the GEF-like protein MON-2, which regulates PE asymmetry and EV release redundantly with sorting nexins independent of the core retromer. Thus, in addition to uncovering redundant intracellular trafficking pathways, our study identifies additional proteins that regulate EV release. This work pinpoints TAT-5 and PE as key regulators of plasma membrane budding, further supporting the model that PE externalization drives ectocytosis.
Keywords: extracellular vesicle; flippase; lipid asymmetry; microvesicle; retromer.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
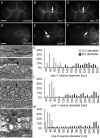
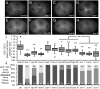
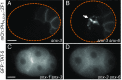
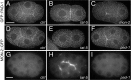
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

