The inhibition of UBC13 expression and blockage of the DNMT1-CHFR-Aurora A pathway contribute to paclitaxel resistance in ovarian cancer
- PMID: 29367628
- PMCID: PMC5833742
- DOI: 10.1038/s41419-017-0137-x
The inhibition of UBC13 expression and blockage of the DNMT1-CHFR-Aurora A pathway contribute to paclitaxel resistance in ovarian cancer
Abstract
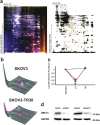
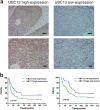
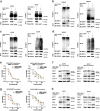
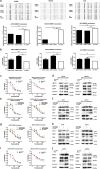
Paclitaxel is widely used as a first-line chemotherapeutic drug for patients with ovarian cancer and other solid cancers, but drug resistance occurs frequently, resulting in ovarian cancer still presenting as the highest lethality among all gynecological tumors. Here, using DIGE quantitative proteomics, we identified UBC13 as down-regulated in paclitaxel-resistant ovarian cancer cells, and it was further revealed by immunohistochemical staining that UBC13 low-expression was associated with poorer prognosis and shorter survival of the patients. Through gene function experiments, we found that paclitaxel exposure induced UBC13 down-regulation, and the enforced change in UBC13 expression altered the sensitivity to paclitaxel. Meanwhile, the reduction of UBC13 increased DNMT1 levels by attenuating its ubiquitination, and the up-regulated DNMT1 enhanced the CHFR promoter DNA methylation levels, leading to a reduction of CHFR expression, and an increased in the levels of Aurora A. Our findings revealed a novel function for UBC13 in regulating paclitaxel sensitivity through a DNMT1-CHFR-Aurora A pathway in ovarian cancer cells. UBC13 could potentially be employed as a therapeutic molecular drug for reversing paclitaxel resistance in ovarian cancer patients.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Aberrant promoter hypermethylation of the CHFR gene in oral squamous cell carcinomas.Oncol Rep. 2009 Nov;22(5):1173-9. doi: 10.3892/or_00000552. Oncol Rep. 2009. PMID: 19787237
-
Alternative efficacy-predicting markers for paclitaxel instead of CHFR in non-small-cell lung cancer.Cancer Biol Ther. 2010 Nov 1;10(9):933-41. doi: 10.4161/cbt.10.9.13320. Epub 2010 Nov 1. Cancer Biol Ther. 2010. PMID: 20855974
-
Poly(ADP‑ribose) polymerase 1/2 inhibitors decrease the ubiquitination of ALC1 mediated by CHFR in breast cancer.Oncol Rep. 2019 Oct;42(4):1467-1474. doi: 10.3892/or.2019.7242. Epub 2019 Jul 18. Oncol Rep. 2019. PMID: 31322269
-
CHFR: a key checkpoint component implicated in a wide range of cancers.Cell Mol Life Sci. 2012 May;69(10):1669-87. doi: 10.1007/s00018-011-0892-2. Epub 2011 Dec 13. Cell Mol Life Sci. 2012. PMID: 22159584 Free PMC article. Review.
-
Emerging evidence for CHFR as a cancer biomarker: from tumor biology to precision medicine.Cancer Metastasis Rev. 2014 Mar;33(1):161-71. doi: 10.1007/s10555-013-9462-4. Cancer Metastasis Rev. 2014. PMID: 24375389 Free PMC article. Review.
Cited by
-
LncRNA SPOCD1-AS from ovarian cancer extracellular vesicles remodels mesothelial cells to promote peritoneal metastasis via interacting with G3BP1.J Exp Clin Cancer Res. 2021 Mar 16;40(1):101. doi: 10.1186/s13046-021-01899-6. J Exp Clin Cancer Res. 2021. PMID: 33726799 Free PMC article.
-
Unraveling role of ubiquitination in drug resistance of gynecological cancer.Am J Cancer Res. 2024 May 15;14(5):2523-2537. doi: 10.62347/WYKZ9784. eCollection 2024. Am J Cancer Res. 2024. PMID: 38859858 Free PMC article. Review.
-
Biocompatible supramolecular pseudorotaxane hydrogels for controllable release of doxorubicin in ovarian cancer SKOV-3 cells.RSC Adv. 2020 Jan 2;10(2):689-697. doi: 10.1039/c9ra08986a. eCollection 2020 Jan 2. RSC Adv. 2020. PMID: 35494427 Free PMC article.
-
Mechanism of Sophorae Flavescentis Radix against ovarian cancer via new pharmacology, molecular docking, and experimental verification.Naunyn Schmiedebergs Arch Pharmacol. 2024 Sep;397(9):6837-6850. doi: 10.1007/s00210-024-03065-z. Epub 2024 Apr 2. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 38561549
-
Research Progress in Prognostic Factors and Biomarkers of Ovarian Cancer.J Cancer. 2021 May 13;12(13):3976-3996. doi: 10.7150/jca.47695. eCollection 2021. J Cancer. 2021. PMID: 34093804 Free PMC article. Review.
References
-
- Ajani JA, et al. Phase II study of taxol in patients with advanced gastric carcinoma. Cancer J. Sci. Am. 1998;4:269–274. - PubMed
-
- Socinski MA, et al. Weekly nab-paclitaxel in combination with carboplatin versus solvent-based paclitaxel plus carboplatin as first-line therapy in patients with advanced non-small-cell lung cancer: final results of a phase III trial. J. Clin. Oncol. 2012;30:2055–2062. doi: 10.1200/JCO.2011.39.5848. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

