Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae
- PMID: 29367657
- PMCID: PMC5783932
- DOI: 10.1038/s41467-017-02576-z
Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae
Abstract
Leprosy is a chronic human disease caused by the yet-uncultured pathogen Mycobacterium leprae. Although readily curable with multidrug therapy (MDT), over 200,000 new cases are still reported annually. Here, we obtain M. leprae genome sequences from DNA extracted directly from patients' skin biopsies using a customized protocol. Comparative and phylogenetic analysis of 154 genomes from 25 countries provides insight into evolution and antimicrobial resistance, uncovering lineages and phylogeographic trends, with the most ancestral strains linked to the Far East. In addition to known MDT-resistance mutations, we detect other mutations associated with antibiotic resistance, and retrace a potential stepwise emergence of extensive drug resistance in the pre-MDT era. Some of the previously undescribed mutations occur in genes that are apparently subject to positive selection, and two of these (ribD, fadD9) are restricted to drug-resistant strains. Finally, nonsense mutations in the nth excision repair gene are associated with greater sequence diversity and drug resistance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
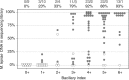
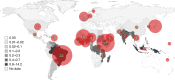


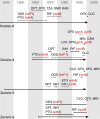
References
-
- World Health Organization. Global leprosy update 2015: time for action, accountability and inclusion. Wkly Epidemiol. Rec. 2016;91:405–420. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

