Parallel derivation of isogenic human primed and naive induced pluripotent stem cells
- PMID: 29367672
- PMCID: PMC5783949
- DOI: 10.1038/s41467-017-02107-w
Parallel derivation of isogenic human primed and naive induced pluripotent stem cells
Abstract
Induced pluripotent stem cells (iPSCs) have considerably impacted human developmental biology and regenerative medicine, notably because they circumvent the use of cells of embryonic origin and offer the potential to generate patient-specific pluripotent stem cells. However, conventional reprogramming protocols produce developmentally advanced, or primed, human iPSCs (hiPSCs), restricting their use to post-implantation human development modeling. Hence, there is a need for hiPSCs resembling preimplantation naive epiblast. Here, we develop a method to generate naive hiPSCs directly from somatic cells, using OKMS overexpression and specific culture conditions, further enabling parallel generation of their isogenic primed counterparts. We benchmark naive hiPSCs against human preimplantation epiblast and reveal remarkable concordance in their transcriptome, dependency on mitochondrial respiration and X-chromosome status. Collectively, our results are essential for the understanding of pluripotency regulation throughout preimplantation development and generate new opportunities for disease modeling and regenerative medicine.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
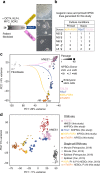
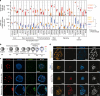
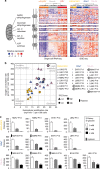


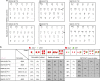
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

