Vertical Light Sheet Enhanced Side-View Imaging for AFM Cell Mechanics Studies
- PMID: 29367675
- PMCID: PMC5784156
- DOI: 10.1038/s41598-018-19791-3
Vertical Light Sheet Enhanced Side-View Imaging for AFM Cell Mechanics Studies
Abstract
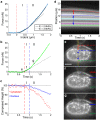
The ability to measure dynamic structural changes within a cell under applied load is essential for developing more accurate models of cell mechanics and mechanotransduction. Atomic force microscopy is a powerful tool for evaluating cell mechanics, but the dominant applied forces and sample strains are in the vertical direction, perpendicular to the imaging plane of standard fluorescence imaging. Here we report on a combined sideways imaging and vertical light sheet illumination system integrated with AFM. Our system enables high frame rate, low background imaging of subcellular structural dynamics in the vertical plane synchronized with AFM force data. Using our system for cell compression measurements, we correlated stiffening features in the force indentation data with onset of nuclear deformation revealed in the imaging data. In adhesion studies we were able to correlate detailed features in the force data during adhesive release events with strain at the membrane and within the nucleus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


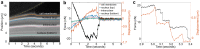

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

