Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production
- PMID: 29367762
- PMCID: PMC6029885
- DOI: 10.1038/s41388-017-0120-0
Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production
Abstract
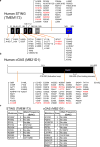
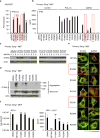
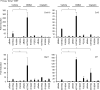
The production of cytokines in response to DNA-damage events may be an important host defense response to help prevent the escape of pre-cancerous cells. The innate immune pathways involved in these events are known to be regulated by cellular molecules such as stimulator of interferon genes (STING), which controls type I interferon and pro-inflammatory cytokine production in response to the presence of microbial DNA or cytosolic DNA that has escaped from the nucleus. STING signaling has been shown to be defective in a variety of cancers, such as colon cancer and melanoma, actions that may enable damaged cells to escape the immunosurveillance system. Here, we report through examination of databases that STING signaling may be commonly suppressed in a greater variety of tumors due to loss-of-function mutation or epigenetic silencing of the STING/cGAS promoter regions. In comparison, RNA activated innate immune pathways controlled by RIG-I/MDA5 were significantly less affected. Examination of reported missense STING variants confirmed that many exhibited a loss-of-function phenotype and could not activate cytokine production following exposure to cytosolic DNA or DNA-damage events. Our data imply that the STING signaling pathway may be recurrently suppressed by a number of mechanisms in a considerable variety of malignant disease and be a requirement for cellular transformation.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

