The DNA methylome of DDR genes and benefit from RT or TMZ in IDH mutant low-grade glioma treated in EORTC 22033
- PMID: 29368212
- PMCID: PMC5978935
- DOI: 10.1007/s00401-018-1810-6
The DNA methylome of DDR genes and benefit from RT or TMZ in IDH mutant low-grade glioma treated in EORTC 22033
Abstract
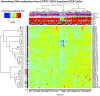
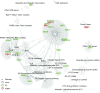
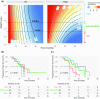
The optimal treatment for patients with low-grade glioma (LGG) WHO grade II remains controversial. Overall survival ranges from 2 to over 15 years depending on molecular and clinical factors. Hence, risk-adjusted treatments are required for optimizing outcome and quality of life. We aim at identifying mechanisms and associated molecular markers predictive for benefit from radiotherapy (RT) or temozolomide (TMZ) in LGG patients treated in the randomized phase III trial EORTC 22033. As candidate biomarkers for these genotoxic treatments, we considered the DNA methylome of 410 DNA damage response (DDR) genes. We first identified 62 functionally relevant CpG sites located in the promoters of 24 DDR genes, using the LGG data from The Cancer Genome Atlas. Then we tested their association with outcome [progression-free survival (PFS)] depending on treatment in 120 LGG patients of EORTC 22033, whose tumors were mutant for isocitrate dehydrogenase 1 or 2 (IDHmt), the molecular hallmark of LGG. The results suggested that seven CpGs of four DDR genes may be predictive for longer PFS in one of the treatment arms that comprised MGMT, MLH3, RAD21, and SMC4. Most interestingly, the two CpGs identified for MGMT are the same, previously selected for the MGMT-STP27 score that is used to determine the methylation status of the MGMT gene. This score was higher in the LGG with 1p/19q codeletion, in this and other independent LGG datasets. It was predictive for PFS in the TMZ, but not in the RT arm of EORTC 22033. The results support the hypothesis that a high score predicts benefit from TMZ treatment for patients with IDHmt LGG, regardless of the 1p/19q status. This MGMT methylation score may identify patients who benefit from first-line treatment with TMZ, to defer RT for long-term preservation of cognitive function and quality of life.
Keywords: DDR genes; DNA methylation; Low-grade glioma; MGMT-STP27; Randomized trial; TMZ.
Conflict of interest statement
Conflict of interest
MJvdB has received grants from Roche and Abbvie, and personal fees from Roche, Abbvie, Merck AG, Novocure, Cavion, Bristol-Myers Squibb, Novartis, and Actelion. BT acknowledges financial support from NCIC-CTG, during the conduct of the study. OC reports grants, personal fees and non-financial support from Roche, and personal fees from Ipsen and AstraZeneca. MJBT reports personal fees from Hoffmann La Roche. W.W. has participated in a speaker’s bureau for and has received research funding from MSD, received research funding from Apogenix, Boehringer Ingelheim, Genentech Roche and Pfizer, has a consultant relationship with BMS, Celldex and Genentech/Roche. AvD reports a patent (US 8,367,347 B2) with royalties paid to Dianova GmbH (Hamburg, Germany). RS received non-financial support from Novocure; his institution received honoraria from Roche, Merck KGaA, MSD, Merck, and Novartis. BGB reports personal fees from Merck Sharp & Dohme (MSD). MEH has received grants from Orion, service fees from Novocure, and has served on advisory board from BMS, and received non-financial support from MDxHealth. The other authors declare that they have no conflict of interest PB, SK, MD; TG, KH-X, EV, AG, RE, FD, AAB, JCR, CM, PF.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Figures


References
-
- Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

