Ubiquitous giants: a plethora of giant viruses found in Brazil and Antarctica
- PMID: 29368617
- PMCID: PMC5784613
- DOI: 10.1186/s12985-018-0930-x
Ubiquitous giants: a plethora of giant viruses found in Brazil and Antarctica
Abstract
Background: Since the discovery of giant viruses infecting amoebae in 2003, many dogmas of virology have been revised and the search for these viruses has been intensified. Over the last few years, several new groups of these viruses have been discovered in various types of samples and environments.In this work, we describe the isolation of 68 giant viruses of amoeba obtained from environmental samples from Brazil and Antarctica.
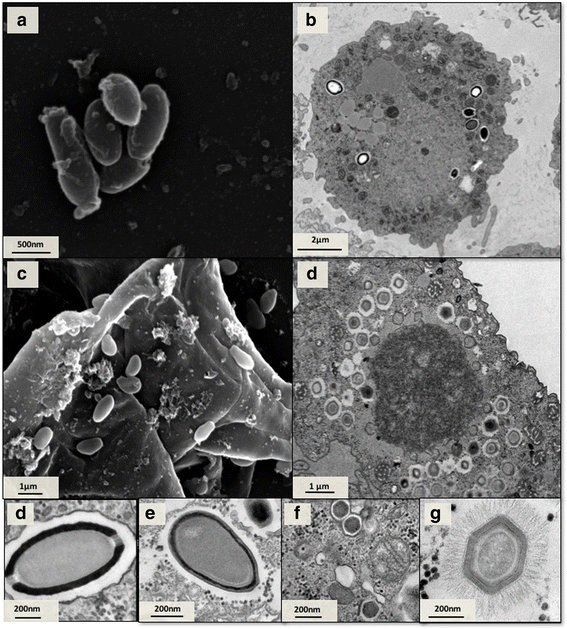
Methods: Isolated viruses were identified by hemacolor staining, PCR assays and electron microscopy (scanning and/or transmission).
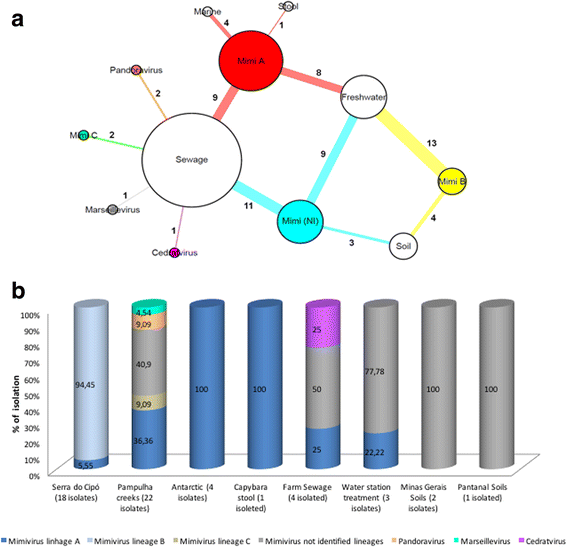
Results: A total of 64 viruses belonging to the Mimiviridae family were isolated (26 from lineage A, 13 from lineage B, 2 from lineage C and 23 from unidentified lineages) from different types of samples, including marine water from Antarctica, thus being the first mimiviruses isolated in this extreme environment to date. Furthermore, a marseillevirus was isolated from sewage samples along with two pandoraviruses and a cedratvirus (the third to be isolated in the world so far).
Conclusions: Considering the different type of samples, we found a higher number of viral groups in sewage samples. Our results reinforce the importance of prospective studies in different environmental samples, therefore improving our comprehension about the circulation anddiversity of these viruses in nature.
Keywords: Antarctica; Brazil; Cedratvirus; Giant viruses; Marseillevirus; Mimivirus; Pandoravirus; Prospection.
Conflict of interest statement
Ethics approval and consent to participate
Two hundred samples of human nasopharyngeal aspiratewere also kindly provided by Laboratório Central do Estado do Rio Grande do Sul (LACEN/RS) (Table 1). These samples were used underapproval of the ethics committee of Universidade Federal de Ciências da Saúde de Porto Alegre (protocol number 1774/12, register 928/12).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

