MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection
- PMID: 29370303
- PMCID: PMC5800688
- DOI: 10.1371/journal.ppat.1006838
MERS-CoV 4b protein interferes with the NF-κB-dependent innate immune response during infection
Abstract
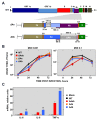
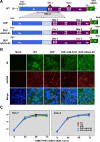
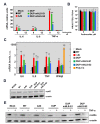
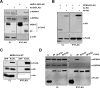
Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel human coronavirus that emerged in 2012, causing severe pneumonia and acute respiratory distress syndrome (ARDS), with a case fatality rate of ~36%. When expressed in isolation, CoV accessory proteins have been shown to interfere with innate antiviral signaling pathways. However, there is limited information on the specific contribution of MERS-CoV accessory protein 4b to the repression of the innate antiviral response in the context of infection. We found that MERS-CoV 4b was required to prevent a robust NF-κB dependent response during infection. In wild-type virus infected cells, 4b localized to the nucleus, while NF-κB was retained in the cytoplasm. In contrast, in the absence of 4b or in the presence of cytoplasmic 4b mutants lacking a nuclear localization signal (NLS), NF-κB was translocated to the nucleus leading to the expression of pro-inflammatory cytokines. This indicates that NF-κB repression required the nuclear import of 4b mediated by a specific NLS. Interestingly, we also found that both in isolation and during infection, 4b interacted with α-karyopherin proteins in an NLS-dependent manner. In particular, 4b had a strong preference for binding karyopherin-α4 (KPNA4), which is known to translocate the NF-κB protein complex into the nucleus. Binding of 4b to KPNA4 during infection inhibited its interaction with NF-κB-p65 subunit. Thereby we propose a model where 4b outcompetes NF-κB for KPNA4 binding and translocation into the nucleus as a mechanism of interference with the NF-κB-mediated innate immune response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

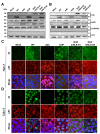
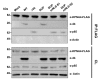

References
-
- Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, et al. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J Virol. 2013;87:5300–4. Epub 2013/03/02. doi: 10.1128/JVI.03496-12 ; PubMed Central PMCID: PMC3624328. - DOI - PMC - PubMed
-
- Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–14. Epub 2013/04/05. doi: 10.1128/JVI.00009-13 ; PubMed Central PMCID: PMC3676115. - DOI - PMC - PubMed
-
- Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, et al. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. MBio. 2013;4:e00611–12. doi: 10.1128/mBio.00611-12 ; PubMed Central PMCID: PMCPMC3573664. - DOI - PMC - PubMed
-
- Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34. doi: 10.1038/nri910 . - DOI - PubMed
-
- DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeño JM, Fernandez-Delgado R, Fett C, et al. Inhibition of NF-kappaB mediated inflammation in severe acute respiratory syndome coronavirus-infected mice increases survival. J Virol. 2014;88:913–24. Epub 2013/11/08. doi: 10.1128/JVI.02576-13 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

