Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae
- PMID: 29371397
- PMCID: PMC5900762
- DOI: 10.1074/jbc.R117.804641
Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae
Abstract
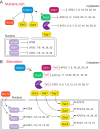
Autophagy is a highly conserved catabolic pathway that is vital for development, cell survival, and the degradation of dysfunctional organelles and potentially toxic aggregates. Dysregulation of autophagy is associated with cancer, neurodegeneration, and lysosomal storage diseases. Accordingly, autophagy is precisely regulated at multiple levels (transcriptional, post-transcriptional, translational, and post-translational) to prevent aberrant activity. Various model organisms are used to study autophagy, but the baker's yeast Saccharomyces cerevisiae continues to be advantageous for genetic and biochemical analysis of non-selective and selective autophagy. In this Minireview, we focus on the cellular mechanisms that regulate autophagy transcriptionally and post-transcriptionally in S. cerevisiae.
Keywords: RNA degradation; autophagy; post-translational modification; transcription; vacuole; yeast.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

