Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines
- PMID: 29371671
- PMCID: PMC5785479
- DOI: 10.1038/s41598-018-19817-w
Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines
Abstract
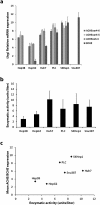
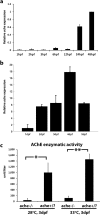
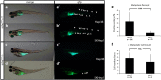
Acetylcholinesterase (AChE), an enzyme responsible for degradation of acetylcholine, has been identified as a prognostic marker in liver cancer. Although in vivo Ache tumorigenicity assays in mouse are present, no established liver cancer xenograft model in zebrafish using an ache mutant background exists. Herein, we developed an embryonic zebrafish xenograft model using epithelial (Hep3B) and mesenchymal (SKHep1) liver cancer cell lines in wild-type and ache sb55 sibling mutant larvae after characterization of cholinesterase expression and activity in cell lines and zebrafish larvae. The comparison of fluorescent signal reflecting tumor size at 3-days post-injection (dpi) revealed an enhanced tumorigenic potential and a reduced migration capacity in cancer cells injected into homozygous ache sb55 mutants when compared with the wild-type. Increased tumor load was confirmed using an ALU based tumor DNA quantification method modified for use in genotyped xenotransplanted zebrafish embryos. Confocal microscopy using the Huh7 cells stably expressing GFP helped identify the distribution of tumor cells in larvae. Our results imply that acetylcholine accumulation in the microenvironment directly or indirectly supports tumor growth in liver cancer. Use of this model system for drug screening studies holds potential in discovering new cholinergic targets for treatment of liver cancers.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

