Draft genome sequence of Streptomyces hyaluromycini MB-PO13T, a hyaluromycin producer
- PMID: 29371910
- PMCID: PMC5765640
- DOI: 10.1186/s40793-017-0286-7
Draft genome sequence of Streptomyces hyaluromycini MB-PO13T, a hyaluromycin producer
Abstract
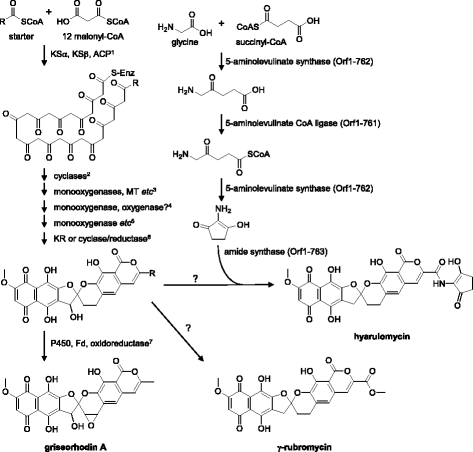
Streptomyces hyaluromycini MB-PO13T (=NBRC 110483T = DSM 100105T) is type strain of the species, which produces a hyaluronidase inhibitor, hyaluromycin. Here, we report the draft genome sequence of this strain together with features of the organism and generation, annotation and analysis of the genome sequence. The 11.5 Mb genome of Streptomyces hyaluromycini MB-PO13T encoded 10,098 putative ORFs, of which 5317 were assigned with COG categories. The genome harbored at least six type I PKS clusters, three type II PKS gene clusters, two type III PKS gene clusters, six NRPS gene clusters, and one hybrid PKS/NRPS gene cluster. The type II PKS gene cluster including 2-amino-3-hydroxycyclopent-2-enone synthetic genes was identified to be responsible for hyaluromycin synthesis. We propose the biosynthetic pathway based on bioinformatic analysis.
Keywords: Biosynthesis; C5N; Polyketide synthase; Rubromycin; Streptomyces.
Conflict of interest statement
The authors declare no competing interest.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Li A, Piel J. A gene cluster from a marine Streptomyces encoding the biosynthesis of the aromatic spiroketal polyketide griseorhodin A. Chem Biol. 2002;9:1017–26. - PubMed
-
- Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int J Syst Evol Microbiol. 1966;16:313–340.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

