Cell-surface marker discovery for lung cancer
- PMID: 29371917
- PMCID: PMC5768334
- DOI: 10.18632/oncotarget.23009
Cell-surface marker discovery for lung cancer
Abstract
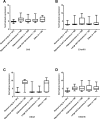
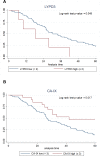
Lung cancer is the leading cause of cancer deaths in the United States. Novel lung cancer targeted therapeutic and molecular imaging agents are needed to improve outcomes and enable personalized care. Since these agents typically cannot cross the plasma membrane while carrying cytotoxic payload or imaging contrast, discovery of cell-surface targets is a necessary initial step. Herein, we report the discovery and characterization of lung cancer cell-surface markers for use in development of targeted agents. To identify putative cell-surface markers, existing microarray gene expression data from patient specimens were analyzed to select markers with differential expression in lung cancer compared to normal lung. Greater than 200 putative cell-surface markers were identified as being overexpressed in lung cancers. Ten cell-surface markers (CA9, CA12, CXorf61, DSG3, FAT2, GPR87, KISS1R, LYPD3, SLC7A11 and TMPRSS4) were selected based on differential mRNA expression in lung tumors vs. non-neoplastic lung samples and other normal tissues, and other considerations involving known biology and targeting moieties. Protein expression was confirmed by immunohistochemistry (IHC) staining and scoring of patient tumor and normal tissue samples. As further validation, marker expression was determined in lung cancer cell lines using microarray data and Kaplan-Meier survival analyses were performed for each of the markers using patient clinical data. High expression for six of the markers (CA9, CA12, CXorf61, GPR87, LYPD3, and SLC7A11) was significantly associated with worse survival. These markers should be useful for the development of novel targeted imaging probes or therapeutics for use in personalized care of lung cancer patients.
Keywords: cell-surface; lung cancer; molecular imaging; target biomarker; targeted therapeutics.
Conflict of interest statement
CONFLICTS OF INTEREST AC and DM are listed as inventors on patent number US20160051704 A1 “Molecular Imaging Probes for Lung Cancer Intraoperative Guidance” that covers the described markers. The other authors declare that they have no conflicts of interest.
Figures







References
-
- Siegel R, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. https://doi.org/10.3322/caac.21387. - DOI - PubMed
-
- American Cancer Society . Cancer Facts & Figures 2017. Atlanta: American Cancer Society; 2017.
-
- Islam S, Walker RC. Advanced imaging (positron emission tomography and magnetic resonance imaging) and image-guided biopsy in initial staging and monitoring of therapy of lung cancer. Cancer J. 2013;19:208–16. https://doi.org/10.1097/PPO.0b013e318295185f. - DOI - PMC - PubMed
-
- Koyama H, Ohno Y, Seki S, Nishio M, Yoshikawa T, Matsumoto S, Sugimura K. Magnetic resonance imaging for lung cancer. J Thorac Imaging. 2013;28:138–50. https://doi.org/10.1097/RTI.0b013e31828d4234. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

