Genome-wide analysis of the human malaria parasite Plasmodium falciparum transcription factor PfNF-YB shows interaction with a CCAAT motif
- PMID: 29371963
- PMCID: PMC5768380
- DOI: 10.18632/oncotarget.23053
Genome-wide analysis of the human malaria parasite Plasmodium falciparum transcription factor PfNF-YB shows interaction with a CCAAT motif
Abstract
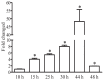
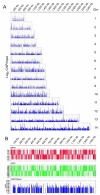
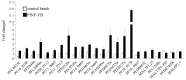
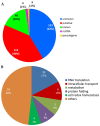
Little is known about transcription factor regulation during the Plasmodium falciparum intraerythrocytic cycle. In order to elucidate the role of the P. falciparum (Pf)NF-YB transcription factor we searched for target genes in the entire genome. PfNF-YB mRNA is highly expressed in late trophozoite and schizont stages relative to the ring stage. In order to determine the candidate genes bound by PfNF-YB a ChIP-on-chip assay was carried out and 297 genes were identified. Ninety nine percent of PfNF-YB binding was to putative promoter regions of protein coding genes of which only 16% comprise proteins of known function. Interestingly, our data reveal that PfNF-YB binding is not exclusively to a canonical CCAAT box motif. PfNF-YB binds to genes coding for proteins implicated in a range of different biological functions, such as replication protein A large subunit (DNA replication), hypoxanthine phosphoribosyltransferase (nucleic acid metabolism) and multidrug resistance protein 2 (intracellular transport).
Keywords: CCAAT-box; Plasmodium falciparum; malaria; signaling; transcription factor.
Conflict of interest statement
COMPETING INTEREST The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
The PfNF-YB transcription factor is a downstream target of melatonin and cAMP signalling in the human malaria parasite Plasmodium falciparum.J Pineal Res. 2013 Mar;54(2):145-53. doi: 10.1111/j.1600-079X.2012.01021.x. Epub 2012 Jul 16. J Pineal Res. 2013. PMID: 22804732
-
Melatonin signaling and its modulation of PfNF-YB transcription factor expression in Plasmodium falciparum.Int J Mol Sci. 2013 Jul 1;14(7):13704-18. doi: 10.3390/ijms140713704. Int J Mol Sci. 2013. PMID: 23839089 Free PMC article. Review.
-
Genome wide in silico analysis of Plasmodium falciparum phosphatome.BMC Genomics. 2014 Nov 25;15:1024. doi: 10.1186/1471-2164-15-1024. BMC Genomics. 2014. PMID: 25425018 Free PMC article.
-
Identification and characterization of NF-YB family genes in tung tree.Mol Genet Genomics. 2015 Dec;290(6):2187-98. doi: 10.1007/s00438-015-1073-z. Epub 2015 Jun 3. Mol Genet Genomics. 2015. PMID: 26037219
-
5' sequence- and chromatin modification-dependent gene expression in Plasmodium falciparum erythrocytic stage.Mol Biochem Parasitol. 2008 Nov;162(1):40-51. doi: 10.1016/j.molbiopara.2008.07.002. Epub 2008 Jul 23. Mol Biochem Parasitol. 2008. PMID: 18692528
Cited by
-
Plasmodium falciparum Development from Gametocyte to Oocyst: Insight from Functional Studies.Microorganisms. 2023 Jul 31;11(8):1966. doi: 10.3390/microorganisms11081966. Microorganisms. 2023. PMID: 37630530 Free PMC article. Review.
-
A nuclear protein, PfMORC confers melatonin dependent synchrony of the human malaria parasite P. falciparum in the asexual stage.Sci Rep. 2021 Jan 21;11(1):2057. doi: 10.1038/s41598-021-81235-2. Sci Rep. 2021. PMID: 33479315 Free PMC article.
-
Target acquired: transcriptional regulators as drug targets for protozoan parasites.Int J Parasitol. 2021 Jul;51(8):599-611. doi: 10.1016/j.ijpara.2020.12.007. Epub 2021 Mar 13. Int J Parasitol. 2021. PMID: 33722681 Free PMC article. Review.
-
Melatonin action in Plasmodium infection: Searching for molecules that modulate the asexual cycle as a strategy to impair the parasite cycle.J Pineal Res. 2021 Jan;70(1):e12700. doi: 10.1111/jpi.12700. Epub 2020 Oct 28. J Pineal Res. 2021. PMID: 33025644 Free PMC article. Review.
-
Evidence of G-Protein-Coupled Receptors (GPCR) in the Parasitic Protozoa Plasmodium falciparum-Sensing the Host Environment and Coupling within Its Molecular Signaling Toolkit.Int J Mol Sci. 2021 Nov 17;22(22):12381. doi: 10.3390/ijms222212381. Int J Mol Sci. 2021. PMID: 34830263 Free PMC article. Review.
References
-
- Garcia CR, de Azevedo MF, Wunderlich G, Budu A, Young JA, Bannister L. Plasmodium in the postgenomic era: new insights into the molecular cell biology of malaria parasites. Int Rev Cell Mol Biol. 2008;266:85–156. - PubMed
-
- Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7:341–54. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

