CoMiniGut-a small volume in vitro colon model for the screening of gut microbial fermentation processes
- PMID: 29372119
- PMCID: PMC5777374
- DOI: 10.7717/peerj.4268
CoMiniGut-a small volume in vitro colon model for the screening of gut microbial fermentation processes
Abstract
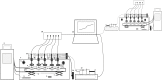
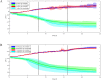
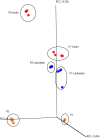
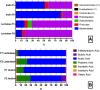
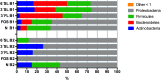
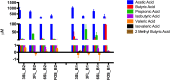
Driven by the growing recognition of the influence of the gut microbiota (GM) on human health and disease, there is a rapidly increasing interest in understanding how dietary components, pharmaceuticals and pre- and probiotics influence GM. In vitro colon models represent an attractive tool for this purpose. With the dual objective of facilitating the investigation of rare and expensive compounds, as well as an increased throughput, we have developed a prototype in vitro parallel gut microbial fermentation screening tool with a working volume of only 5 ml consisting of five parallel reactor units that can be expanded with multiples of five to increase throughput. This allows e.g., the investigation of interpersonal variations in gut microbial dynamics and the acquisition of larger data sets with enhanced statistical inference. The functionality of the in vitro colon model, Copenhagen MiniGut (CoMiniGut) was first demonstrated in experiments with two common prebiotics using the oligosaccharide inulin and the disaccharide lactulose at 1% (w/v). We then investigated fermentation of the scarce and expensive human milk oligosaccharides (HMOs) 3-Fucosyllactose, 3-Sialyllactose, 6-Sialyllactose and the more common Fructooligosaccharide in fermentations with infant gut microbial communities. Investigations of microbial community composition dynamics in the CoMiniGut reactors by MiSeq-based 16S rRNA gene amplicon high throughput sequencing showed excellent experimental reproducibility and allowed us to extract significant differences in gut microbial composition after 24 h of fermentation for all investigated substrates and fecal donors. Furthermore, short chain fatty acids (SCFAs) were quantified for all treatments and donors. Fermentations with inulin and lactulose showed that inulin leads to a microbiota dominated by obligate anaerobes, with high relative abundance of Bacteroidetes, while the more easily fermented lactulose leads to higher relative abundance of Proteobacteria. The subsequent study on the influence of HMOs on two infant GM communities, revealed the strongest bifidogenic effect for 3'SL for both infants. Inter-individual differences of infant GM, especially with regards to the occurrence of Bacteroidetes and differences in bifidobacterial species composition, correlated with varying degrees of HMO utilization foremost of 6'SL and 3'FL, indicating species and strain related differences in HMO utilization which was also reflected in SCFAs concentrations, with 3'SL and 6'SL resulting in significantly higher butyrate production compared to 3'FL. In conclusion, the increased throughput of CoMiniGut strengthens experimental conclusions through elimination of statistical interferences originating from low number of repetitions. Its small working volume moreover allows the investigation of rare and expensive bioactives.
Keywords: 16S rRNA gene; Bifidogenic effect; Gut microbiome; Human milk oligosaccharide; In vitro colon model; Short chain fatty acids.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
References
-
- Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, Sears MR, Becker AB, Scott JA, Kozyrskyj AL. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Canadian Medical Association Journal. 2013;185:385–394. doi: 10.1503/cmaj.121189. - DOI - PMC - PubMed
-
- Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. The Journal of Nutrition. 2006;136:2127–2130. - PubMed
-
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

