Divide and Control: Comparison of Split and Switch Hybridization Sensors
- PMID: 29372178
- PMCID: PMC5777618
- DOI: 10.1002/slct.201701179
Divide and Control: Comparison of Split and Switch Hybridization Sensors
Abstract
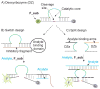
Hybridization probes have been intensively used for nucleic acid analysis in medicine, forensics and fundamental research. Instantaneous hybridization probes (IHPs) enable signalling immediately after binding to a targeted DNA or RNA sequences without the need to isolate the probe-target complex (e. g. by gel electrophoresis). The two most common strategies for IHP design are conformational switches and split approach. A conformational switch changes its conformation and produces signal upon hybridization to a target. Split approach uses two (or more) strands that independently or semi independently bind the target and produce an output signal only if all components associate. Here, we compared the performance of split vs switch designs for deoxyribozyme (Dz) hybridization probes under optimal conditions for each of them. The split design was represented by binary Dz (BiDz) probes; while catalytic molecular beacon (CMB) probes represented the switch design. It was found that BiDz were significantly more selective than CMBs in recognition of single base substitution. CMBs produced high background signal when operated at 55°C. An important advantage of BiDz over CMB is more straightforward design and simplicity of assay optimization.
Keywords: Catalytic molecular beacons; Deoxyribozyme sensors; fluorescent sensors; instantaneous hybridization probes; single nucleotide polymorphisms; split hybridization probes.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
