Developmental profiling of microRNAs in the human embryonic inner ear
- PMID: 29373586
- PMCID: PMC5786302
- DOI: 10.1371/journal.pone.0191452
Developmental profiling of microRNAs in the human embryonic inner ear
Abstract
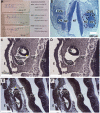
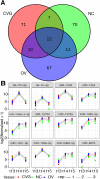
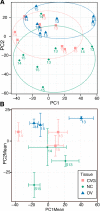
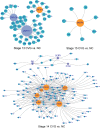
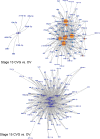
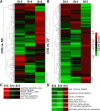
Due to the extreme inaccessibility of fetal human inner ear tissue, defining of the microRNAs (miRNAs) that regulate development of the inner ear has relied on animal tissue. In the present study, we performed the first miRNA sequencing of otic precursors in human specimens. Using HTG miRNA Whole Transcriptome assays, we examined miRNA expression in the cochleovestibular ganglion (CVG), neural crest (NC), and otic vesicle (OV) from paraffin embedded (FFPE) human specimens in the Carnegie developmental stages 13-15. We found that in human embryonic tissues, there are different patterns of miRNA expression in the CVG, NC and OV. In particular, members of the miR-183 family (miR-96, miR-182, and miR-183) are differentially expressed in the CVG compared to NC and OV at Carnegie developmental stage 13. We further identified transcription factors that are differentially targeted in the CVG compared to the other tissues from stages 13-15, and we performed gene set enrichment analyses to determine differentially regulated pathways that are relevant to CVG development in humans. These findings not only provide insight into the mechanisms governing the development of the human inner ear, but also identify potential signaling pathways for promoting regeneration of the spiral ganglion and other components of the inner ear.
Conflict of interest statement
Figures


References
-
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91: 817–887. doi: 10.1152/physrev.00006.2010 MICRORNAS - DOI - PubMed
-
- Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. Nature Publishing Group; 2014;15: 565–76. doi: 10.1038/nrm3854 - DOI - PMC - PubMed
-
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup G a. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111: 95–104. doi: 10.1016/j.brainres.2006.07.006 - DOI - PubMed
-
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. Elsevier Inc.; 2009;328: 328–341. doi: 10.1016/j.ydbio.2009.01.037 - DOI - PMC - PubMed
-
- Friedman LM, Dror A a, Avraham KB. Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol. 2007;51: 609–31. doi: 10.1387/ijdb.072365lf - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

