Methanogens Are Major Contributors to Nitrogen Fixation in Soils of the Florida Everglades
- PMID: 29374038
- PMCID: PMC5861825
- DOI: 10.1128/AEM.02222-17
Methanogens Are Major Contributors to Nitrogen Fixation in Soils of the Florida Everglades
Abstract
The objective of this study was to investigate the interaction of the nitrogen (N) cycle with methane production in the Florida Everglades, a large freshwater wetland. This study provides an initial analysis of the distribution and expression of N-cycling genes in Water Conservation Area 2A (WCA-2A), a section of the marsh that underwent phosphorus (P) loading for many years due to runoff from upstream agricultural activities. The elevated P resulted in increased primary productivity and an N limitation in P-enriched areas. Results from quantitative real-time PCR (qPCR) analyses indicated that the N cycle in WCA-2A was dominated by nifH and nirK/S, with an increasing trend in copy numbers in P-impacted sites. Many nifH sequences (6 to 44% of the total) and nifH transcript sequences (2 to 49%) clustered with the methanogenic Euryarchaeota, in stark contrast to the proportion of core gene sequences representing Archaea (≤0.27% of SSU rRNA genes) for the WCA-2A microbiota. Notably, archaeal nifH gene transcripts were detected at all sites and comprised a significant proportion of total nifH transcripts obtained from the unimpacted site, indicating that methanogens are actively fixing N2 Laboratory incubations with soils taken from WCA-2A produced nifH transcripts with the production of methane from H2 plus CO2 and acetate as electron donors and carbon sources. Methanogenic N2 fixation is likely to be an important, although largely unrecognized, route through which fixed nitrogen enters the anoxic soils of the Everglades and may have significant relevance regarding methane production in wetlands.IMPORTANCE Wetlands are the most important natural sources of the greenhouse gas methane, and much of that methane emanates from (sub)tropical peatlands. Primary productivity in these peatlands is frequently limited by the availability of nitrogen or phosphorus; however, the response to nutrient limitations of microbial communities that control biogeochemical cycling critical to ecosystem function may be complex and may be associated with a range of processes, including methane production. We show that many, if not most, of the methanogens in the peatlands of the Florida Everglades possess the nifH gene and actively express it for N2 fixation coupled with methanogenesis. These findings indicate that archaeal N2 fixation would play crucial role in methane emissions and overall N cycle in subtropical wetlands suffering N limitation.
Keywords: Everglades; methanogenesis; methanogens; nifH gene; nitrogen cycle; nitrogen fixation.
Copyright © 2018 American Society for Microbiology.
Figures

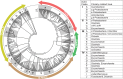



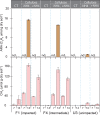
References
-
- van Amstel A. 2012. Methane. A review. Integr Environ Sci 9:5–30. doi: 10.1080/1943815X.2012.694892. - DOI
-
- DeBusk WF, Reddy KR, Koch MS, Wang Y. 1994. Spatial distribution of soil nutrients in a northern Everglades marsh: Water Conservation Area 2A. Soil Sci Soc Am J 58:543–552. doi: 10.2136/sssaj1994.03615995005800020042x. - DOI
-
- Craft CB, Richardson CJ. 2008. Soil characteristics of the Everglades peatland, p 59–72. In Richardson CJ. (ed), The Everglades experiments: lessons for ecosystem restoration. Springer, New York, NY.
-
- Corstanje R, Reddy KR, Prenger JP, Newman S, Ogram A. 2007. Soil microbial eco-physiological response to nutrient enrichment in a sub-tropical wetland. Ecol Indic 7:277–289. doi: 10.1016/j.ecolind.2006.02.002. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

