Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes
- PMID: 29374454
- PMCID: PMC5787289
- DOI: 10.1186/s12870-018-1237-1
Resistance to Plum Pox Virus (PPV) in apricot (Prunus armeniaca L.) is associated with down-regulation of two MATHd genes
Abstract
Background: Plum pox virus (PPV), causing Sharka disease, is one of the main limiting factors for Prunus production worldwide. In apricot (Prunus armeniaca L.) the major PPV resistance locus (PPVres), comprising ~ 196 kb, has been mapped to the upper part of linkage group 1. Within the PPVres, 68 genomic variants linked in coupling to PPV resistance were identified within 23 predicted transcripts according to peach genome annotation. Taking into account the predicted functions inferred from sequence homology, some members of a cluster of meprin and TRAF-C homology domain (MATHd)-containing genes were pointed as PPV resistance candidate genes.
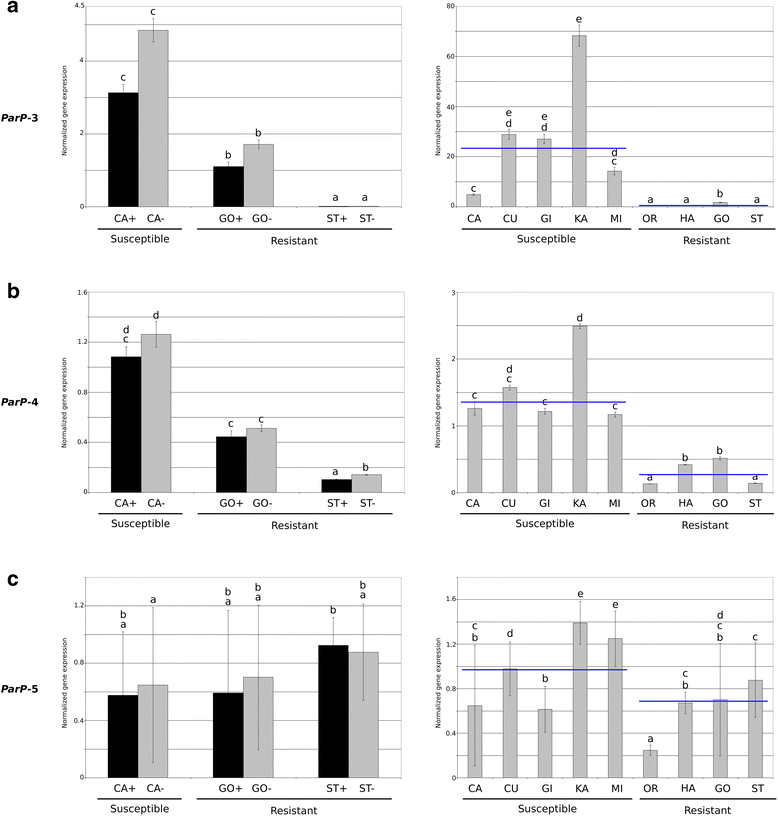
Results: Here, we have characterized the global apricot transcriptome response to PPV-D infection identifying six PPVres locus genes (ParP-1 to ParP-6) differentially expressed in resistant/susceptible cultivars. Two of them (ParP-3 and ParP-4), that encode MATHd proteins, appear clearly down-regulated in resistant cultivars, as confirmed by qRT-PCR. Concurrently, variant calling was performed using whole-genome sequencing data of 24 apricot cultivars (10 PPV-resistant and 14 PPV-susceptible) and 2 wild relatives (PPV-susceptible). ParP-3 and ParP-4, named as Prunus armeniaca PPVres MATHd-containing genes (ParPMC), are the only 2 genes having allelic variants linked in coupling to PPV resistance. ParPMC1 has 1 nsSNP, while ParPMC2 has 15 variants, including a 5-bp deletion within the second exon that produces a frameshift mutation. ParPMC1 and ParPMC2 are adjacent and highly homologous (87.5% identity) suggesting they are paralogs originated from a tandem duplication. Cultivars carrying the ParPMC2 resistant (mutated) allele show lack of expression in both ParPMC2 and especially ParPMC1.
Conclusions: Accordingly, we hypothesize that ParPMC2 is a pseudogene that mediates down-regulation of its functional paralog ParPMC1 by silencing. As a whole, results strongly support ParPMC1 and/or ParPMC2 as host susceptibility genes required for PPV infection which silencing may confer PPV resistance trait. This finding may facilitate resistance breeding by marker-assisted selection and pave the way for gene edition approaches in Prunus.
Keywords: Apricot; MATHd; PPV resistance; Plum Pox virus; Potyvirus; Prunus; Silencing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they do not have competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Asensio M. El virus de la sharka (plum pox virus): caracterización, diagnóstico y detección mediante anticuerpos monoclonales específicos. PhD dissertation. Spain: University of Valencia; 1996. p. 193.
-
- Atanasoff D. Plum Pox. A New Virus Disease. In: Yearbook Faculty Agricultural University 1932/1933, Sofia, Bulgaria. 1933;11:49–69.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

