Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties
- PMID: 29374777
- PMCID: PMC6132922
- DOI: 10.1007/s00535-018-1437-3
Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties
Abstract
Background: Intestinal stem cells (ISCs) play indispensable roles in the maintenance of homeostasis, and also in the regeneration of the damaged intestinal epithelia. However, whether the inflammatory environment of Crohn's disease (CD) affects properties of resident small intestinal stem cells remain uncertain.
Methods: CD patient-derived small intestinal organoids were established from enteroscopic biopsy specimens taken from active lesions (aCD-SIO), or from mucosa under remission (rCD-SIO). Expression of ISC-marker genes in those organoids was examined by immunohistochemistry, and also by microfluid-based single-cell multiplex gene expression analysis. The ISC-specific function of organoid cells was evaluated using a single-cell organoid reformation assay.
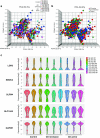
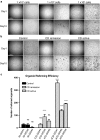
Results: ISC-marker genes, OLFM4 and SLC12A2, were expressed by an increased number of small intestinal epithelial cells in the active lesion of CD. aCD-SIOs, rCD-SIOs or those of non-IBD controls (NI-SIOs) were successfully established from 9 patients. Immunohistochemistry showed a comparable level of OLFM4 and SLC12A2 expression in all organoids. Single-cell gene expression data of 12 ISC-markers were acquired from a total of 1215 cells. t-distributed stochastic neighbor embedding analysis identified clusters of candidate ISCs, and also revealed a distinct expression pattern of SMOC2 and LGR5 in ISC-cluster classified cells derived from aCD-SIOs. Single-cell organoid reformation assays showed significantly higher reformation efficiency by the cells of the aCD-SIOs compared with that of cells from NI-SIOs.
Conclusions: aCD-SIOs harbor ISCs with modified marker expression profiles, and also with high organoid reformation ability. Results suggest modification of small intestinal stem cell properties by unidentified factors in the inflammatory environment of CD.
Keywords: Crohn’s disease; Intestinal organoids; Intestinal stem cell; Single cell analysis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

