Tomato Apical Leaf Curl Virus: A Novel, Monopartite Geminivirus Detected in Tomatoes in Argentina
- PMID: 29375528
- PMCID: PMC5770407
- DOI: 10.3389/fmicb.2017.02665
Tomato Apical Leaf Curl Virus: A Novel, Monopartite Geminivirus Detected in Tomatoes in Argentina
Abstract
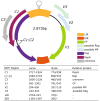
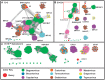
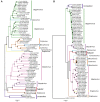
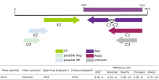
Plant viruses that are members of the Geminiviridae family have circular single-stranded DNA (ssDNA) genome and are responsible for major crop diseases worldwide. We have identified and characterized a novel monopartite geminivirus infecting tomato in Argentina. The full-length genome was cloned and sequenced. The genome-wide pairwise identity calculation that resulted in a maximum of 63% identity with all of other known geminiviruses indicated that it is a new geminivirus species. Biolistic infected plants presented interveinal yellowing, apical leaf curling and extreme root hypotrophy. Thus, the name proposed for this species is tomato apical leaf curl virus (ToALCV). The phylogenetic inferences suggested different evolutionary relationships for the replication-associated protein (Rep) and the coat protein (CP). Besides, the sequence similarity network (SSN) protein analyses showed that the complementary-sense gene products (RepA, Rep and C3) are similar to capulavirus while the viron-sense gene products (CP, MP and V3) are similar to topocuvirus, curtovirus and becurtovirus. Based on the data presented, ToALCV genome appears to have "modular organization" supported by its recombination origin. Analyses of the specificity-determining positions (SDPs) of the CP of geminiviruses defined nine subgroups that include geminiviruses that share the same type of insect vector. Our sequences were clustered with the sequences of topocuvirus, whose vector is the treehopper, Micrutalis malleifera. Also, a set of the highest scored amino acid residues was predicted for the CP, which could determine differences in virus transmission specificity. We predict that a treehopper could be the vector of ToALCV, but transmission assays need to be performed to confirm this. Given everything we demonstrate in this paper, ToALCV can be considered a type member of a new putative genus of the Geminiviridae family.
Keywords: Argentina; ToALCV; coat protein; recombination; sequence similarity network; specificity-determining positions; tomato; treehopper.
Figures


References
-
- Andrade E. C., Manhani G. G., Alfenas P. F., Calegario R. F., Fontes E. P., Zerbini F. M. (2006). Tomato yellow spot virus, a tomato-infecting begomovirus from Brazil with a closer relationship to viruses from Sida sp., forms pseudorecombinants with begomoviruses from tomato but not from Sida. J. Gen. Virol. 87 3687–3696. 10.1099/vir.0.82279-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

