Transcriptional Modulation of Penicillin-Binding Protein 1b, Outer Membrane Protein P2 and Efflux Pump (AcrAB-TolC) during Heat Stress Is Correlated to Enhanced Bactericidal Action of Imipenem on Non-typeable Haemophilus influenzae
- PMID: 29375536
- PMCID: PMC5770572
- DOI: 10.3389/fmicb.2017.02676
Transcriptional Modulation of Penicillin-Binding Protein 1b, Outer Membrane Protein P2 and Efflux Pump (AcrAB-TolC) during Heat Stress Is Correlated to Enhanced Bactericidal Action of Imipenem on Non-typeable Haemophilus influenzae
Abstract
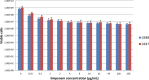
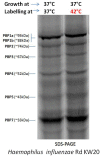
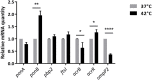
Objective: The purpose of the present study was to investigate the penicillin binding proteins (PBPs), drug influx and efflux modulations during heat stress and their effects on the bactericidal action of imipenem on non-typeable Haemophilus influenzae (NTHi). Methods: The two NTHi clinical isolates (GE47 and GE88, imipenem MICs by E-test > 32 μg/mL) examined in this study were collected at Geneva University Hospitals. The imipenem killing activity was assessed after incubation of the NTHi strains at either 37 or 42°C for 3 h with increasing concentrations of imipenem. The detection of PBPs was carried out by Bocillin-FL. Global transcriptional changes were monitored by RNA-seq after pre-incubation of bacterial cells at either 37 or 42°C, and the expression levels of relevant target genes were confirmed by qRT-PCR. Results: Quantitation of NTHi viable cells after incubation with 0.25 μg/mL of imipenem for 3 h revealed more than a twofold decrease in GE47 and GE88 viable cells at 42°C as compared to 37°C. Transcriptome analysis showed that under heat stress conditions, there were 141 differentially expressed genes with a | log2(fold change)| > 1, including 67 up-regulated and 74 down-regulated genes. The expression levels of ponB (encoding PBP1b) and acrR (regulator of AcrAB-TolC efflux pump) were significantly increased at 42°C. In contrast, the transcript levels of ompP2 (encoding the outer membrane protein P2) and acrB gene (encoding AcrB) were significantly lower under heat stress condition. Conclusion: This study shows that the transcriptional modulation of ponB, ompP2, acrR, and acrB in the heat stress response is correlated to enhanced antimicrobial effects of imipenem on non-typeable H. influenzae.
Keywords: AcrAB-TolC; Bocillin-FL; NTHi; OmpP2; PBP1b; RNA-seq; heat stress.
Figures




Similar articles
-
Imipenem heteroresistance in nontypeable Haemophilus influenzae is linked to a combination of altered PBP3, slow drug influx and direct efflux regulation.Clin Microbiol Infect. 2017 Feb;23(2):118.e9-118.e19. doi: 10.1016/j.cmi.2016.10.009. Epub 2016 Oct 15. Clin Microbiol Infect. 2017. PMID: 27756711
-
Role of the AcrAB-TolC efflux pump in determining susceptibility of Haemophilus influenzae to the novel peptide deformylase inhibitor LBM415.Antimicrob Agents Chemother. 2005 Aug;49(8):3129-35. doi: 10.1128/AAC.49.8.3129-3135.2005. Antimicrob Agents Chemother. 2005. PMID: 16048914 Free PMC article.
-
First characterization of heterogeneous resistance to imipenem in invasive nontypeable Haemophilus influenzae isolates.Antimicrob Agents Chemother. 2007 Sep;51(9):3155-61. doi: 10.1128/AAC.00335-07. Epub 2007 Jul 9. Antimicrob Agents Chemother. 2007. PMID: 17620383 Free PMC article.
-
Identification of AcrAB-TolC Efflux Pump Genes and Detection of Mutation in Efflux Repressor AcrR from Omeprazole Responsive Multidrug-Resistant Escherichia coli Isolates Causing Urinary Tract Infections.Microbiol Insights. 2019 Dec 4;12:1178636119889629. doi: 10.1177/1178636119889629. eCollection 2019. Microbiol Insights. 2019. PMID: 31839709 Free PMC article.
-
What the pediatrician should know about non-typeable Haemophilus influenzae.J Infect. 2015 Jun;71 Suppl 1:S10-4. doi: 10.1016/j.jinf.2015.04.014. Epub 2015 Apr 25. J Infect. 2015. PMID: 25917803 Review.
Cited by
-
Inactivation of farR Causes High Rhodomyrtone Resistance and Increased Pathogenicity in Staphylococcus aureus.Front Microbiol. 2019 May 28;10:1157. doi: 10.3389/fmicb.2019.01157. eCollection 2019. Front Microbiol. 2019. PMID: 31191485 Free PMC article.
-
Imipenem heteroresistance but not tolerance in Haemophilus influenzae during chronic lung infection associated with chronic obstructive pulmonary disease.Front Microbiol. 2023 Dec 20;14:1253623. doi: 10.3389/fmicb.2023.1253623. eCollection 2023. Front Microbiol. 2023. PMID: 38179447 Free PMC article.
-
Multiple interspecies recombination events documented by whole-genome sequencing in multidrug-resistant Haemophilus influenzae clinical isolates.Access Microbiol. 2024 Feb 12;6(2):000649.v3. doi: 10.1099/acmi.0.000649.v3. eCollection 2024. Access Microbiol. 2024. PMID: 38482359 Free PMC article.
-
Learning from -omics strategies applied to uncover Haemophilus influenzae host-pathogen interactions: Current status and perspectives.Comput Struct Biotechnol J. 2021 May 15;19:3042-3050. doi: 10.1016/j.csbj.2021.05.026. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34136102 Free PMC article. Review.
-
Revealing the Response Mechanism of Pediococcus pentosaceus Under Acid and Alcohol Stresses via a Combined Transcriptomic and Metabolomic Analysis.Foods. 2025 Jul 7;14(13):2400. doi: 10.3390/foods14132400. Foods. 2025. PMID: 40647152 Free PMC article.
References
-
- Cherkaoui A., Diene S. M., Renzoni A., Emonet S., Renzi G., Francois P., et al. (2016). Imipenem heteroresistance in nontypeable Haemophilus influenzae is linked to a combination of altered PBP3, slow drug influx and direct efflux regulation. Clin. Microbiol. Infect. 23 118.e9–118.e19. 10.1016/j.cmi.2016.10.009 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

