Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study
- PMID: 29375539
- PMCID: PMC5770402
- DOI: 10.3389/fmicb.2017.02699
Preliminary Comparison of Oral and Intestinal Human Microbiota in Patients with Colorectal Cancer: A Pilot Study
Abstract
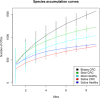
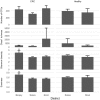
In this study Next-Generation Sequencing (NGS) was used to analyze and compare human microbiota from three different compartments, i.e., saliva, feces, and cancer tissue (CT), of a selected cohort of 10 Italian patients with colorectal cancer (CRC) vs. 10 healthy controls (saliva and feces). Furthermore, the Fusobacterium nucleatum abundance in the same body site was investigated through real-time quantitative polymerase chain reaction (qPCR) to assess the association with CRC. Differences in bacterial composition, F. nucleatum abundance in healthy controls vs. CRC patients, and the association of F. nucleatum with clinical parameters were observed. Taxonomic analysis based on 16S rRNA gene, revealed the presence of three main bacterial phyla, which includes about 80% of reads: Firmicutes (39.18%), Bacteroidetes (30.36%), and Proteobacteria (10.65%). The results highlighted the presence of different bacterial compositions; in particular, the fecal samples of CRC patients seemed to be enriched with Bacteroidetes, whereas in the fecal samples of healthy controls Firmicutes were one of the major phyla detected though these differences were not statistically significant. The CT samples showed the highest alpha diversity values. These results emphasize a different taxonomic composition of feces from CRC compared to healthy controls. Despite the low number of samples included in the study, these results suggest the importance of microbiota in the CRC progression and could pave the way to the development of therapeutic interventions and novel microbial-related diagnostic tools in CRC patients.
Keywords: Fusobacterium nucleatum; colorectal cancer; gut microbiota; oral microbiota; quantitative polymerase chain reaction; taxonomic analysis.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

