Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection
- PMID: 29375588
- PMCID: PMC5770625
- DOI: 10.3389/fpls.2017.02016
Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection
Abstract
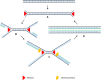
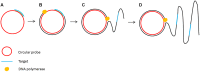
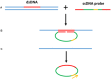
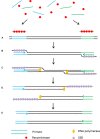
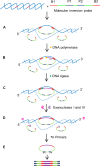
Diagnostic technologies for the detection of plant pathogens with point-of-care capability and high multiplexing ability are an essential tool in the fight to reduce the large agricultural production losses caused by plant diseases. The main desirable characteristics for such diagnostic assays are high specificity, sensitivity, reproducibility, quickness, cost efficiency and high-throughput multiplex detection capability. This article describes and discusses various DNA-based point-of care diagnostic methods for applications in plant disease detection. Polymerase chain reaction (PCR) is the most common DNA amplification technology used for detecting various plant and animal pathogens. However, subsequent to PCR based assays, several types of nucleic acid amplification technologies have been developed to achieve higher sensitivity, rapid detection as well as suitable for field applications such as loop-mediated isothermal amplification, helicase-dependent amplification, rolling circle amplification, recombinase polymerase amplification, and molecular inversion probe. The principle behind these technologies has been thoroughly discussed in several review papers; herein we emphasize the application of these technologies to detect plant pathogens by outlining the advantages and disadvantages of each technology in detail.
Keywords: HDA; LAMP; RCA; RPA; isothermal amplification; point-of-care diagnostic.
Figures

References
-
- Agrios G. N. (2005). Plant Pathology. Amsterdam: Elsevier Academic Press.
-
- Ahmadi S., Almasi M. A., Fatehi F., Struik P. C., Moradi A. (2013). Visual detection of potato leafroll virus by one-step reverse transcription loop-mediated isothermal amplification of DNA with hydroxynaphthol blue dye. J. Phytopathol. 161 120–124. 10.1111/jph.12037 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

