No Time to Waste: Transcriptome Study Reveals that Drought Tolerance in Barley May Be Attributed to Stressed-Like Expression Patterns that Exist before the Occurrence of Stress
- PMID: 29375595
- PMCID: PMC5767312
- DOI: 10.3389/fpls.2017.02212
No Time to Waste: Transcriptome Study Reveals that Drought Tolerance in Barley May Be Attributed to Stressed-Like Expression Patterns that Exist before the Occurrence of Stress
Abstract
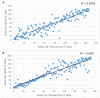
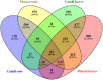
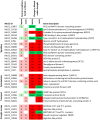
Plant survival in adverse environmental conditions requires a substantial change in the metabolism, which is reflected by the extensive transcriptome rebuilding upon the occurrence of the stress. Therefore, transcriptomic studies offer an insight into the mechanisms of plant stress responses. Here, we present the results of global gene expression profiling of roots and leaves of two barley genotypes with contrasting ability to cope with drought stress. Our analysis suggests that drought tolerance results from a certain level of transcription of stress-influenced genes that is present even before the onset of drought. Genes that predispose the plant to better drought survival play a role in the regulatory network of gene expression, including several transcription factors, translation regulators and structural components of ribosomes. An important group of genes is involved in signaling mechanisms, with significant contribution of hormone signaling pathways and an interplay between ABA, auxin, ethylene and brassinosteroid homeostasis. Signal transduction in a drought tolerant genotype may be more efficient through the expression of genes required for environmental sensing that are active already during normal water availability and are related to actin filaments and LIM domain proteins, which may function as osmotic biosensors. Better survival of drought may also be attributed to more effective processes of energy generation and more efficient chloroplasts biogenesis. Interestingly, our data suggest that several genes involved in a photosynthesis process are required for the establishment of effective drought response not only in leaves, but also in roots of barley. Thus, we propose a hypothesis that root plastids may turn into the anti-oxidative centers protecting root macromolecules from oxidative damage during drought stress. Specific genes and their potential role in building up a drought-tolerant barley phenotype is extensively discussed with special emphasis on processes that take place in barley roots. When possible, the interconnections between particular factors are emphasized to draw a broader picture of the molecular mechanisms of drought tolerance in barley.
Keywords: barley; drought tolerance; root system; stress; transcriptomics.
Figures




References
-
- Aimar D., Calafat M., Andrade A. M., Carassay L., Abdala G. I., Molas M. L. (2011). Drought Tolerance and Stress Hormones: From Model Organisms to Forage Crops. Intech: From Model Organisms to Forage Crops. Available online at: https://www.intechopen.com/books/plants-and-environment/drought-toleranc...
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

