Mapping the Chromatin State Dynamics in Myoblasts
- PMID: 29376142
- PMCID: PMC5786272
- DOI: 10.1016/j.genrep.2016.01.003
Mapping the Chromatin State Dynamics in Myoblasts
Abstract
Background: Genome-wide mapping reveals chromatin landscapes unique to cell states. Histone marks of regulatory genes involved in cell specification and organ development provide a powerful tool to map regulatory sequences. H3K4me3 marks promoter regions; H3K27me3 marks repressed regions, and Pol II presence indicates active transcription. The presence of both H3K4me3 and H3K27me3 characterize poised sequences, a common characteristic of genes involved in pattern formation during organogenesis.
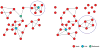
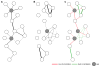
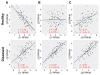
Results: We used genome-wide profiling for H3K27me3, H3K4me3, and Pol II to map chromatin states in mouse embryonic day 12 forelimbs in wild type (control) and Pitx2-null mutant mice. We compared these data with previous gene expression studies from forelimb Lbx1+ migratory myoblasts and correlated Pitx2-dependent expression profiles and chromatin states. During forelimb development, several lineages including myoblast, osteoblast, neurons, angioblasts etc., require synchronized growth to form a functional limb. We identified 125 genes in the developing forelimb that are Pitx2-dependent. Genes involved in muscle specification and cytoskeleton architecture were positively regulated, while genes involved in axonal path finding were poised.
Conclusion: Our results have established histone modification profiles as a useful tool for identifying gene regulatory states in muscle development, and identified the role of Pitx2 in extending the time of myoblast progression, promoting formation of sarcomeric structures, and suppressing attachment of neuronal axons.
Keywords: chromatin; homeobox; muscle development; myogenesis.
Figures
References
-
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. - PubMed
-
- Amthor H, Christ B, Patel K. A molecular mechanism enabling continuous embryonic muscle growth - a balance between proliferation and differentiation. Development. 1999;126:1041–1053. - PubMed
-
- Arber S, Hunter JJ, Ross J, Hongo M, Sansig G, Borg J, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. CELL. 1997;88:393–403. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, … Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
