The genome of the Hi5 germ cell line from Trichoplusia ni, an agricultural pest and novel model for small RNA biology
- PMID: 29376823
- PMCID: PMC5844692
- DOI: 10.7554/eLife.31628
The genome of the Hi5 germ cell line from Trichoplusia ni, an agricultural pest and novel model for small RNA biology
Abstract
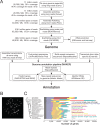
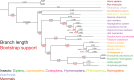
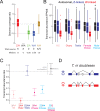
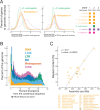
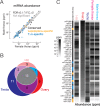
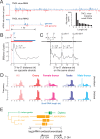
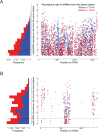
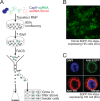
We report a draft assembly of the genome of Hi5 cells from the lepidopteran insect pest, Trichoplusia ni, assigning 90.6% of bases to one of 28 chromosomes and predicting 14,037 protein-coding genes. Chemoreception and detoxification gene families reveal T. ni-specific gene expansions that may explain its widespread distribution and rapid adaptation to insecticides. Transcriptome and small RNA data from thorax, ovary, testis, and the germline-derived Hi5 cell line show distinct expression profiles for 295 microRNA- and >393 piRNA-producing loci, as well as 39 genes encoding small RNA pathway proteins. Nearly all of the W chromosome is devoted to piRNA production, and T. ni siRNAs are not 2´-O-methylated. To enable use of Hi5 cells as a model system, we have established genome editing and single-cell cloning protocols. The T. ni genome provides insights into pest control and allows Hi5 cells to become a new tool for studying small RNAs ex vivo.
Keywords: Lepidoptera; T. ni; Tricoplusia ni; chromosomes; evolutionary biology; genes; genomics; insecticide resistance; piRNAs; siRNAs.
© 2018, Fu et al.
Conflict of interest statement
YF, YY, HZ, GF, JW, KQ, ZW, PZ No competing interests declared
Figures










References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

