Partial DNA-guided Cas9 enables genome editing with reduced off-target activity
- PMID: 29377001
- PMCID: PMC5902734
- DOI: 10.1038/nchembio.2559
Partial DNA-guided Cas9 enables genome editing with reduced off-target activity
Abstract
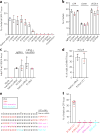
CRISPR-Cas9 is a versatile RNA-guided genome editing tool. Here we demonstrate that partial replacement of RNA nucleotides with DNA nucleotides in CRISPR RNA (crRNA) enables efficient gene editing in human cells. This strategy of partial DNA replacement retains on-target activity when used with both crRNA and sgRNA, as well as with multiple guide sequences. Partial DNA replacement also works for crRNA of Cpf1, another CRISPR system. We find that partial DNA replacement in the guide sequence significantly reduces off-target genome editing through focused analysis of off-target cleavage, measurement of mismatch tolerance and genome-wide profiling of off-target sites. Using the structure of the Cas9-sgRNA complex as a guide, the majority of the 3' end of crRNA can be replaced with DNA nucleotide, and the 5 - and 3'-DNA-replaced crRNA enables efficient genome editing. Cas9 guided by a DNA-RNA chimera may provide a generalized strategy to reduce both the cost and the off-target genome editing in human cells.
Conflict of interest statement
The authors declare competing financial interests: details accompany the online version of the paper.
Figures

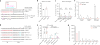

References
-
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

