A robust gene signature for the prediction of early relapse in stage I-III colon cancer
- PMID: 29377588
- PMCID: PMC5891048
- DOI: 10.1002/1878-0261.12175
A robust gene signature for the prediction of early relapse in stage I-III colon cancer
Abstract
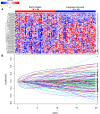
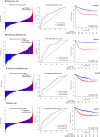
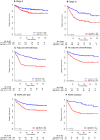
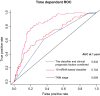
Colon cancer patients experiencing early relapse consistently exhibited poor survival. The aim of our study was to develop an mRNA signature that can help to detect early relapse cases in stage I-III colon cancer. Public microarray datasets of stage I-III colon cancer samples were extracted from the Gene Expression Omnibus database. Propensity score matching analysis was performed between patients in the early relapse group and the long-term survival group from GSE39582 discovery series (N = 386), and patients were 1 : 1 matched. Global mRNA expression changes were then analyzed between the paired groups to identify the differentially expressed genes. Lasso Cox regression modeling analysis was conducted for the selection of prognostic mRNA. Fifteen mRNA were finally identified to build an early relapse classifier. With specific risk score formula, patients were classified into a high-risk group and a low-risk group. Relapse-free survival was significantly different between the two groups in every series, including discovery [hazard ratio (HR): 2.547, 95% confidence interval (CI): 1.708-3.797, P < 0.001)], internal validation (HR: 5.146, 95% CI: 1.968-13.457, P < 0.001), and external validation (HR: 1.977, 95% CI: 1.295-3.021, P < 0.001) sets of patients. Time-dependent receiver-operating characteristic at 1 year suggested more prognostic accuracy of the classifier [area under curve (AUC = 0.703)] than the American Joint Commission on Cancer tumor-node-metastasis staging system (AUC = 0.659) in all 951 patients. In conclusion, we developed a robust mRNA signature that can effectively classify colon cancer patients into groups with low and high risks of early relapse. This mRNA signature may help select high-risk colon cancer patients who require more aggressive therapeutic intervention.
Keywords: Gene Expression Omnibus database; colon cancer; early relapse; mRNA signature; propensity score.
© 2018 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures
References
-
- Alexopoulou DK, Papadopoulos IN and Scorilas A (2013) Clinical significance of kallikrein‐related peptidase (KLK10) mRNA expression in colorectal cancer. Clin Biochem 46, 1453–1461. - PubMed
-
- Becker H (1995) Surgery of colorectal carcinoma. Praxis 84, 1371–1372. - PubMed
-
- Berghella AM, Contasta I, Pellegrini P, Del Beato T and Adorno D (2002) Peripheral blood immunological parameters for use as markers of pre‐invasive to invasive colorectal cancer. Cancer Biother Radiopharm 17, 43–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

