Reactome graph database: Efficient access to complex pathway data
- PMID: 29377902
- PMCID: PMC5805351
- DOI: 10.1371/journal.pcbi.1005968
Reactome graph database: Efficient access to complex pathway data
Abstract
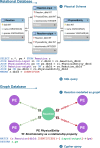
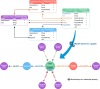
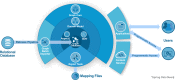
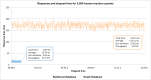
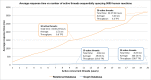
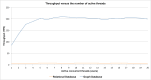
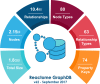
Reactome is a free, open-source, open-data, curated and peer-reviewed knowledgebase of biomolecular pathways. One of its main priorities is to provide easy and efficient access to its high quality curated data. At present, biological pathway databases typically store their contents in relational databases. This limits access efficiency because there are performance issues associated with queries traversing highly interconnected data. The same data in a graph database can be queried more efficiently. Here we present the rationale behind the adoption of a graph database (Neo4j) as well as the new ContentService (REST API) that provides access to these data. The Neo4j graph database and its query language, Cypher, provide efficient access to the complex Reactome data model, facilitating easy traversal and knowledge discovery. The adoption of this technology greatly improved query efficiency, reducing the average query time by 93%. The web service built on top of the graph database provides programmatic access to Reactome data by object oriented queries, but also supports more complex queries that take advantage of the new underlying graph-based data storage. By adopting graph database technology we are providing a high performance pathway data resource to the community. The Reactome graph database use case shows the power of NoSQL database engines for complex biological data types.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016; 44:D481–7. doi: 10.1093/nar/gkv1351 - DOI - PMC - PubMed
-
- Van Bruggen R. Learning Neo4j. Birmingham: Packt Publishing Ltd.; 2014
-
- Vukotic A, Watt N, Abedrabbo T, Fox D, Partner J. Neo4j in Action. 1st ed. Shelter Island, NY: Manning Publications; 2014.
-
- Sedgewick R, Wayne K. Algorithms. 4th ed. Addison-Wesley; 2011. pp. 566–596.
-
- Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, Croft D, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007; 8: R39 doi: 10.1186/gb-2007-8-3-r39 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

