ProtDataTherm: A database for thermostability analysis and engineering of proteins
- PMID: 29377907
- PMCID: PMC5788348
- DOI: 10.1371/journal.pone.0191222
ProtDataTherm: A database for thermostability analysis and engineering of proteins
Abstract
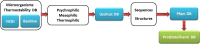
Protein thermostability engineering is a powerful tool to improve resistance of proteins against high temperatures and thereafter broaden their applications. For efficient protein thermostability engineering, different thermostability-classified data sources including sequences and 3D structures are needed for different protein families. However, no data source is available providing such data easily. It is the first release of ProtDataTherm database for analysis and engineering of protein thermostability which contains more than 14 million protein sequences categorized based on their thermal stability and protein family. This database contains data needed for better understanding protein thermostability and stability engineering. Providing categorized protein sequences and structures as psychrophilic, mesophilic and thermophilic makes this database useful for the development of new tools in protein stability prediction. This database is available at http://profiles.bs.ipm.ir/softwares/protdatatherm. As a proof of concept, the thermostability that improves mutations were suggested for one sample protein belonging to one of protein families with more than 20 mesophilic and thermophilic sequences and with known experimentally measured ΔT of mutations available within ProTherm database.
Conflict of interest statement
Figures
Similar articles
-
Applications of Protein Thermodynamic Database for Understanding Protein Mutant Stability and Designing Stable Mutants.Methods Mol Biol. 2016;1415:71-89. doi: 10.1007/978-1-4939-3572-7_4. Methods Mol Biol. 2016. PMID: 27115628
-
Structure Based Thermostability Prediction Models for Protein Single Point Mutations with Machine Learning Tools.PLoS One. 2015 Sep 11;10(9):e0138022. doi: 10.1371/journal.pone.0138022. eCollection 2015. PLoS One. 2015. PMID: 26361227 Free PMC article.
-
Amino acid coupling patterns in thermophilic proteins.Proteins. 2005 Apr 1;59(1):58-63. doi: 10.1002/prot.20386. Proteins. 2005. PMID: 15688447
-
Engineering proteins for thermostability through rigidifying flexible sites.Biotechnol Adv. 2014 Mar-Apr;32(2):308-15. doi: 10.1016/j.biotechadv.2013.10.012. Epub 2013 Nov 6. Biotechnol Adv. 2014. PMID: 24211474 Review.
-
[Prediction of protein thermostability from their primary structure: the current state and development factors].Biomed Khim. 2017 Mar;63(2):124-131. doi: 10.18097/PBMC20176302124. Biomed Khim. 2017. PMID: 28414283 Review. Russian.
Cited by
-
Systematic evaluation of computational tools to predict the effects of mutations on protein stability in the absence of experimental structures.Brief Bioinform. 2022 Mar 10;23(2):bbac025. doi: 10.1093/bib/bbac025. Brief Bioinform. 2022. PMID: 35189634 Free PMC article.
-
TemBERTure: advancing protein thermostability prediction with deep learning and attention mechanisms.Bioinform Adv. 2024 Jul 13;4(1):vbae103. doi: 10.1093/bioadv/vbae103. eCollection 2024. Bioinform Adv. 2024. PMID: 39040220 Free PMC article.
-
Multidisciplinary involvement and potential of thermophiles.Folia Microbiol (Praha). 2019 May;64(3):389-406. doi: 10.1007/s12223-018-0662-8. Epub 2018 Nov 1. Folia Microbiol (Praha). 2019. PMID: 30386965 Review.
-
Role of simple descriptors and applicability domain in predicting change in protein thermostability.PLoS One. 2018 Sep 7;13(9):e0203819. doi: 10.1371/journal.pone.0203819. eCollection 2018. PLoS One. 2018. PMID: 30192891 Free PMC article.
-
A Method for Prediction of Thermophilic Protein Based on Reduced Amino Acids and Mixed Features.Front Bioeng Biotechnol. 2020 May 5;8:285. doi: 10.3389/fbioe.2020.00285. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32432088 Free PMC article.
References
-
- Elleuche S, Schäfers C, Blank S, Schröder C, Antranikian G. Exploration of extremophiles for high temperature biotechnological processes. Current Opinion in Microbiology. 2015;25:113–9. doi: 10.1016/j.mib.2015.05.011 - DOI - PubMed
-
- Gromiha MM, Oobatake M, Sarai A. Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophysical Chemistry. 1999;82(1):51–67. - PubMed
-
- Pezeshgi Modarres H, Dorokhov BD, Popov VO, Ravin NV, Skryabin KG, Dal Peraro M. Understanding and engineering thermostability in DNA ligase from Thermococcus sp. 1519. Biochemistry. 2015;54(19):3076–85. doi: 10.1021/bi501227b - DOI - PubMed
-
- Panigrahi P, Sule M, Ghanate A, Ramasamy S, Suresh C. Engineering proteins for thermostability with iRDP web server. PloS One. 2015;10(10):e0139486 doi: 10.1371/journal.pone.0139486 - DOI - PMC - PubMed
-
- Chaparro-Riggers JF, Polizzi KM, Bommarius AS. Better library design: data-driven protein engineering. Biotechnology Journal. 2007;2(2):180–91. doi: 10.1002/biot.200600170 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials