Partial growth hormone insensitivity and dysregulatory immune disease associated with de novo germline activating STAT3 mutations
- PMID: 29378236
- PMCID: PMC6143347
- DOI: 10.1016/j.mce.2018.01.016
Partial growth hormone insensitivity and dysregulatory immune disease associated with de novo germline activating STAT3 mutations
Abstract
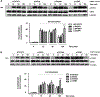
Germinal heterozygous activating STAT3 mutations represent a novel monogenic defect associated with multi-organ autoimmune disease and, in some cases, severe growth retardation. By using whole-exome sequencing, we identified two novel STAT3 mutations, p.E616del and p.C426R, in two unrelated pediatric patients with IGF-I deficiency and immune dysregulation. The functional analyses showed that both variants were gain-of-function (GOF), although they were not constitutively phosphorylated. They presented differences in their dephosphorylation kinetics and transcriptional activities under interleukin-6 stimulation. Both variants increased their transcriptional activities in response to growth hormone (GH) treatment. Nonetheless, STAT5b transcriptional activity was diminished in the presence of STAT3 GOF variants, suggesting a disruptive role of STAT3 GOF variants in the GH signaling pathway. This study highlights the broad clinical spectrum of patients presenting activating STAT3 mutations and explores the underlying molecular pathway responsible for this condition, suggesting that different mutations may drive increased activity by slightly different mechanisms.
Keywords: Activating mutations; Growth hormone insensitivity; IGF-I deficiency; Immune dysregulation; STAT3.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interest
The authors declare no conflict of interest.
Figures





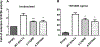
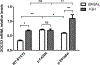
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

