Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic evolutionary history
- PMID: 29378510
- PMCID: PMC5789663
- DOI: 10.1186/s12864-018-4487-2
Transcriptomic profiling of Burkholderia phymatum STM815, Cupriavidus taiwanensis LMG19424 and Rhizobium mesoamericanum STM3625 in response to Mimosa pudica root exudates illuminates the molecular basis of their nodulation competitiveness and symbiotic evolutionary history
Abstract
Background: Rhizobial symbionts belong to the classes Alphaproteobacteria and Betaproteobacteria (called "alpha" and "beta"-rhizobia). Most knowledge on the genetic basis of symbiosis is based on model strains belonging to alpha-rhizobia. Mimosa pudica is a legume that offers an excellent opportunity to study the adaptation toward symbiotic nitrogen fixation in beta-rhizobia compared to alpha-rhizobia. In a previous study (Melkonian et al., Environ Microbiol 16:2099-111, 2014) we described the symbiotic competitiveness of M. pudica symbionts belonging to Burkholderia, Cupriavidus and Rhizobium species.
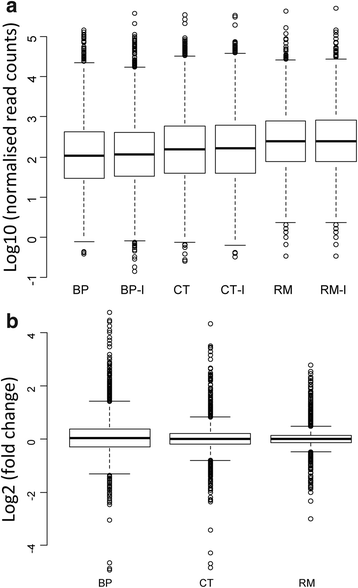
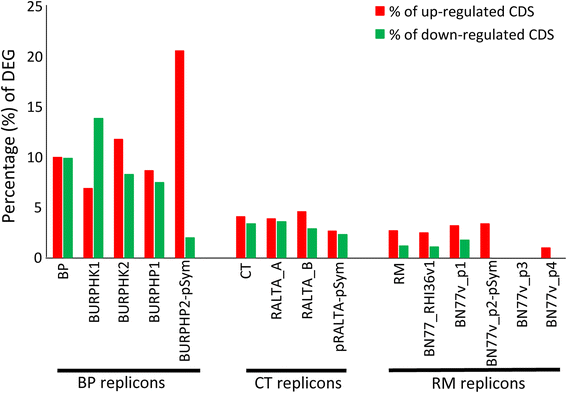
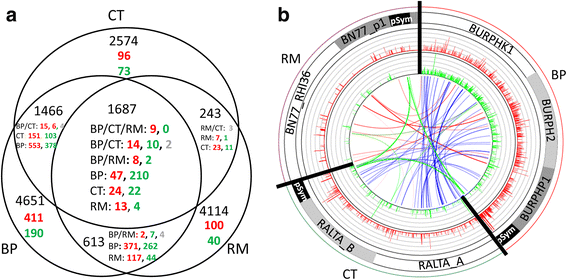
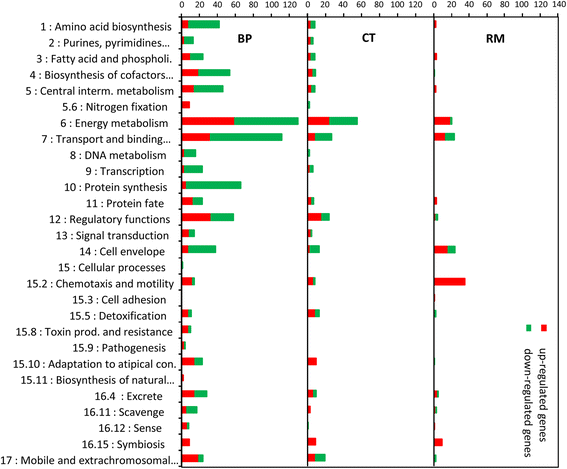
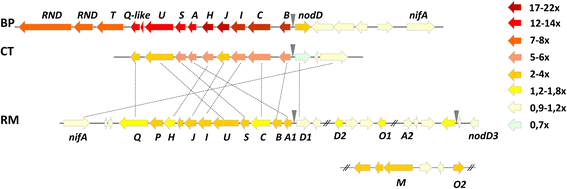
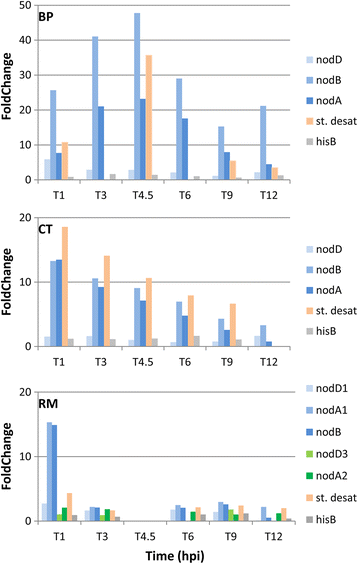
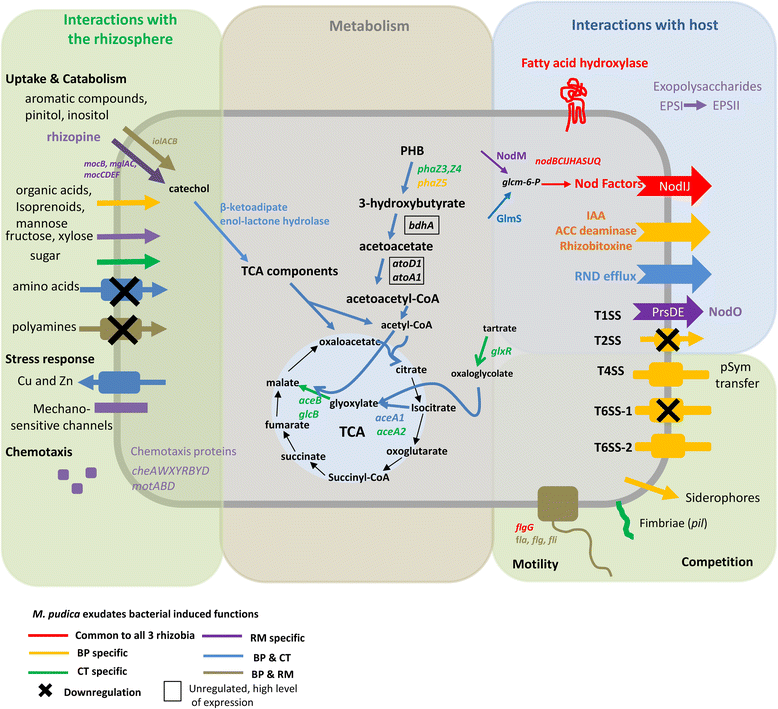
Results: In this article we present a comparative analysis of the transcriptomes (by RNAseq) of B. phymatum STM815 (BP), C. taiwanensis LMG19424 (CT) and R. mesoamericanum STM3625 (RM) in conditions mimicking the early steps of symbiosis (i.e. perception of root exudates). BP exhibited the strongest transcriptome shift both quantitatively and qualitatively, which mirrors its high competitiveness in the early steps of symbiosis and its ancient evolutionary history as a symbiont, while CT had a minimal response which correlates with its status as a younger symbiont (probably via acquisition of symbiotic genes from a Burkholderia ancestor) and RM had a typical response of Alphaproteobacterial rhizospheric bacteria. Interestingly, the upregulation of nodulation genes was the only common response among the three strains; the exception was an up-regulated gene encoding a putative fatty acid hydroxylase, which appears to be a novel symbiotic gene specific to Mimosa symbionts.
Conclusion: The transcriptional response to root exudates was correlated to each strain nodulation competitiveness, with Burkholderia phymatum appearing as the best specialised symbiont of Mimosa pudica.
Keywords: Alpha-rhizobia; Beta-rhizobia; RNA-seq; Symbiosis; Transcriptome.
Conflict of interest statement
Ethics approval
This study has not directly involved humans or animals. Plants used in this study were cultivated from seeds purchased at B&T World Seeds company (
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype.Environ Microbiol. 2014 Jul;16(7):2099-111. doi: 10.1111/1462-2920.12286. Epub 2013 Oct 17. Environ Microbiol. 2014. PMID: 24131520
-
Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta.New Phytol. 2007;173(1):168-80. doi: 10.1111/j.1469-8137.2006.01894.x. New Phytol. 2007. PMID: 17176403
-
Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils.FEMS Microbiol Ecol. 2012 Sep;81(3):618-35. doi: 10.1111/j.1574-6941.2012.01393.x. Epub 2012 May 14. FEMS Microbiol Ecol. 2012. PMID: 22512707
-
Experimental Evolution of Legume Symbionts: What Have We Learnt?Genes (Basel). 2020 Mar 23;11(3):339. doi: 10.3390/genes11030339. Genes (Basel). 2020. PMID: 32210028 Free PMC article. Review.
-
Specificity in Legume-Rhizobia Symbioses.Int J Mol Sci. 2017 Mar 26;18(4):705. doi: 10.3390/ijms18040705. Int J Mol Sci. 2017. PMID: 28346361 Free PMC article. Review.
Cited by
-
Root Exudates Alter the Expression of Diverse Metabolic, Transport, Regulatory, and Stress Response Genes in Rhizosphere Pseudomonas.Front Microbiol. 2021 Apr 14;12:651282. doi: 10.3389/fmicb.2021.651282. eCollection 2021. Front Microbiol. 2021. PMID: 33936009 Free PMC article.
-
Discovery of a novel filamentous prophage in the genome of the Mimosa pudica microsymbiont Cupriavidus taiwanensis STM 6018.Front Microbiol. 2023 Feb 28;14:1082107. doi: 10.3389/fmicb.2023.1082107. eCollection 2023. Front Microbiol. 2023. PMID: 36925474 Free PMC article.
-
Scent of a Symbiont: The Personalized Genetic Relationships of Rhizobium-Plant Interaction.Int J Mol Sci. 2022 Mar 20;23(6):3358. doi: 10.3390/ijms23063358. Int J Mol Sci. 2022. PMID: 35328782 Free PMC article. Review.
-
Functional Genomics Approaches to Studying Symbioses between Legumes and Nitrogen-Fixing Rhizobia.High Throughput. 2018 May 18;7(2):15. doi: 10.3390/ht7020015. High Throughput. 2018. PMID: 29783718 Free PMC article. Review.
-
Inoculation of Mimosa Pudica with Paraburkholderia phymatum Results in Changes to the Rhizoplane Microbial Community Structure.Microbes Environ. 2021;36(1):ME20153. doi: 10.1264/jsme2.ME20153. Microbes Environ. 2021. PMID: 33716243 Free PMC article.
References
-
- Dénarié J, Debellé F, Promé JC. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem. 1996;65:503–535. - PubMed
-
- Debellé F, Moulin L, Mangin B, Dénarié J, Boivin C. Nod genes and nod signals and the evolution of the Rhizobium legume symbiosis. Acta Biochim Pol. 2001;48:359–365. - PubMed
-
- Moulin L, Munive A, Dreyfus B, Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature. 2001;411:948–950. - PubMed
-
- Gyaneshwar P, Hirsch AM, Moulin L, Chen W-M, Elliott GN, Bontemps C, et al. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol Plant-Microbe Interact. 2011;24:1276–1288. - PubMed
-
- Moulin L, James EK, Klonowska A, Miana de Faria S, Simon MF. Phylogeny, diversity, geographical distribution, and host range of legume-nodulating betaproteobacteria : what is the role of plant taxonomy ? In: de Bruijn FJ. Biol. nitrogen Fixat. Chichester: Wiley; 2015. p. 177–190.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

