Detection and genotyping of Helicobacter pylori in saliva versus stool samples from asymptomatic individuals in Northeastern Thailand reveals intra-host tissue-specific H. pylori subtypes
- PMID: 29378521
- PMCID: PMC5789744
- DOI: 10.1186/s12866-018-1150-7
Detection and genotyping of Helicobacter pylori in saliva versus stool samples from asymptomatic individuals in Northeastern Thailand reveals intra-host tissue-specific H. pylori subtypes
Abstract
Background: Two-thirds of the world's population is thought to be infected by Helicobacter pylori. Although most people infected with H. pylori are asymptomatic, this pathogen is associated with several gastric pathologies including cancer. The risk factors for colonization are still unclear and the genetic diversity within individual hosts has never been clearly investigated.
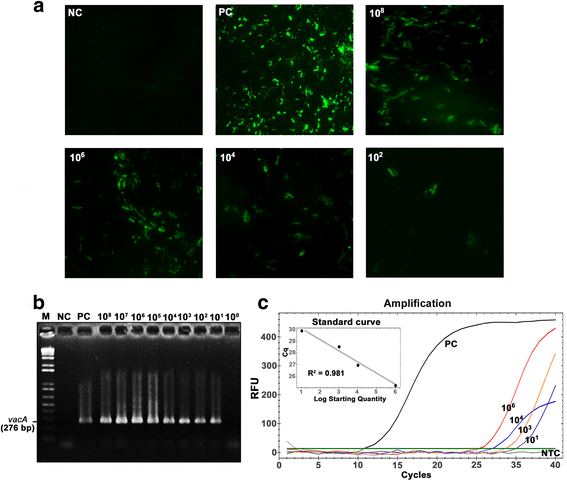
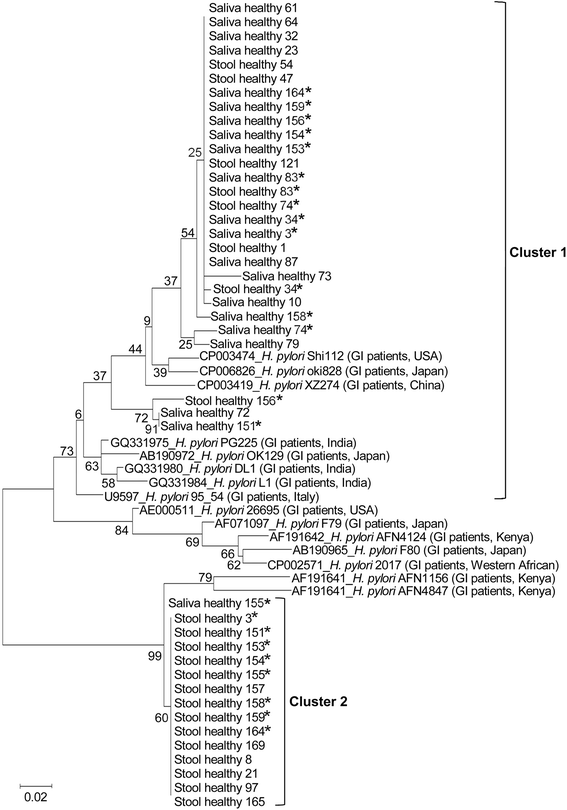
Result: This study determined the prevalence of, and explored risk factors for, H. pylori infection directly from paired saliva (n = 110) and stool (n = 110) samples from asymptomatic persons in Northeast Thailand. Samples were subjected to indirect immunofluorescence assay (IFA), 16S rRNA-based real-time PCR and vacA-based semi-nested PCR. Partial vacA gene sequences of H. pylori were compared between saliva and stool samples. The overall prevalence of H. pylori infection in our asymptomatic study population was 64%. Age, gender, occupation and frequency of brushing teeth were not found to be associated with H. pylori colonization. The vacA gene was successfully sequenced from both saliva and stool samples of 12 individuals. For seven of these individuals, saliva and stool sequences fell into different clusters on a phylogenetic tree, indicating intra-host genetic variation of H. pylori.
Conclusion: This study reports a high prevalence of H. pylori infection in asymptomatic persons in this region of Thailand and demonstrates that genotypes (vacA gene sequences) of H. pylori may differ between the oral cavity and intestinal tract.
Keywords: Genetic diversity; Genotyping; Helicobacter pylori; Saliva; Stool; vacA.
Conflict of interest statement
Ethics approval and consent to participate
Written ethical consents were received from participants. The study was approved by Institutional Human Ethics Committee of Khon Kaen University (HE571489).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Pounder RE, Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995;9(Suppl 2):33–39. - PubMed
-
- Leelawat K, Suksumek N, Leelawat S, Lek-Uthai U. Detection of VacA gene specific for Helicobactor pylori in hepatocellular carcinoma and cholangiocarcinoma specimens of Thai patients. Southeast Asian J Trop Med Public Health. 2007;38(5):881–885. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

