Visualizing transcription factor dynamics in living cells
- PMID: 29378780
- PMCID: PMC5881510
- DOI: 10.1083/jcb.201710038
Visualizing transcription factor dynamics in living cells
Abstract
The assembly of sequence-specific enhancer-binding transcription factors (TFs) at cis-regulatory elements in the genome has long been regarded as the fundamental mechanism driving cell type-specific gene expression. However, despite extensive biochemical, genetic, and genomic studies in the past three decades, our understanding of molecular mechanisms underlying enhancer-mediated gene regulation remains incomplete. Recent advances in imaging technologies now enable direct visualization of TF-driven regulatory events and transcriptional activities at the single-cell, single-molecule level. The ability to observe the remarkably dynamic behavior of individual TFs in live cells at high spatiotemporal resolution has begun to provide novel mechanistic insights and promises new advances in deciphering causal-functional relationships of TF targeting, genome organization, and gene activation. In this review, we review current transcription imaging techniques and summarize converging results from various lines of research that may instigate a revision of models to describe key features of eukaryotic gene regulation.
© 2018 Liu and Tjian.
Figures

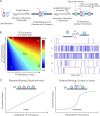
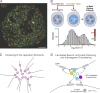

References
-
- Ball D.A., Mehta G.D., Salomon-Kent R., Mazza D., Morisaki T., Mueller F., McNally J.G., and Karpova T.S.. 2016. Single molecule tracking of Ace1p in Saccharomyces cerevisiae defines a characteristic residence time for non-specific interactions of transcription factors with chromatin. Nucleic Acids Res. 44:e160. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

