Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function
- PMID: 29378810
- PMCID: PMC5788517
- DOI: 10.1534/genetics.117.300386
Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function
Abstract
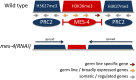
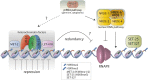
Chromatin is organized and compacted in the nucleus through the association of histones and other proteins, which together control genomic activity. Two broad types of chromatin can be distinguished: euchromatin, which is generally transcriptionally active, and heterochromatin, which is repressed. Here we examine the current state of our understanding of repressed chromatin in Caenorhabditis elegans, focusing on roles of histone modifications associated with repression, such as methylation of histone H3 lysine 9 (H3K9me2/3) or the Polycomb Repressive Complex 2 (MES-2/3/6)-deposited modification H3K27me3, and on proteins that recognize these modifications. Proteins involved in chromatin repression are important for development, and have demonstrated roles in nuclear organization, repetitive element silencing, genome integrity, and the regulation of euchromatin. Additionally, chromatin factors participate in repression with small RNA pathways. Recent findings shed light on heterochromatin function and regulation in C. elegans, and should inform our understanding of repressed chromatin in other animals.
Keywords: C. elegans; H3K27me; H3K9me; WormBook; chromatin; heterochromatin; histone methylation.
Copyright © 2018 by the Genetics Society of America.
Figures



References
-
- Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., et al. , 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. - PubMed
-
- Aguilera A., Garcia-Muse T., 2012. R loops: from transcription byproducts to threats to genome stability. Mol. Cell 46: 115–124. - PubMed
-
- Albertson D. G., Thomson J. N., 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86: 409–428. - PubMed
-
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

