Effects of Distal Mutations on the Structure, Dynamics and Catalysis of Human Monoacylglycerol Lipase
- PMID: 29379013
- PMCID: PMC5789057
- DOI: 10.1038/s41598-017-19135-7
Effects of Distal Mutations on the Structure, Dynamics and Catalysis of Human Monoacylglycerol Lipase
Abstract
An understanding of how conformational dynamics modulates function and catalysis of human monoacylglycerol lipase (hMGL), an important pharmaceutical target, can facilitate the development of novel ligands with potential therapeutic value. Here, we report the discovery and characterization of an allosteric, regulatory hMGL site comprised of residues Trp-289 and Leu-232 that reside over 18 Å away from the catalytic triad. These residues were identified as critical mediators of long-range communication and as important contributors to the integrity of the hMGL structure. Nonconservative replacements of Trp-289 or Leu-232 triggered concerted motions of structurally distinct regions with a significant conformational shift toward inactive states and dramatic loss in catalytic efficiency of the enzyme. Using a multimethod approach, we show that the dynamically relevant Trp-289 and Leu-232 residues serve as communication hubs within an allosteric protein network that controls signal propagation to the active site, and thus, regulates active-inactive interconversion of hMGL. Our findings provide new insights into the mechanism of allosteric regulation of lipase activity, in general, and may provide alternative drug design possibilities.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



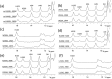


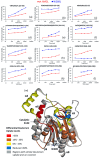
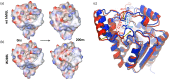
Similar articles
-
Conformational gating, dynamics and allostery in human monoacylglycerol lipase.Sci Rep. 2020 Oct 28;10(1):18531. doi: 10.1038/s41598-020-75497-5. Sci Rep. 2020. PMID: 33116203 Free PMC article.
-
Identification by nuclear magnetic resonance spectroscopy of an active-site hydrogen-bond network in human monoacylglycerol lipase (hMGL): implications for hMGL dynamics, pharmacological inhibition, and catalytic mechanism.Mol Biosyst. 2010 Aug;6(8):1381-8. doi: 10.1039/c004515b. Epub 2010 May 12. Mol Biosyst. 2010. PMID: 20464001 Free PMC article.
-
Endocannabinoid enzyme engineering: soluble human thio-monoacylglycerol lipase (sol-S-hMGL).ACS Chem Neurosci. 2012 May 16;3(5):393-9. doi: 10.1021/cn3000263. Epub 2012 Mar 20. ACS Chem Neurosci. 2012. PMID: 22860208 Free PMC article.
-
Allosteric regulation and catalysis emerge via a common route.Nat Chem Biol. 2008 Aug;4(8):474-82. doi: 10.1038/nchembio.98. Nat Chem Biol. 2008. PMID: 18641628 Review.
-
Monoglyceride lipase: Structure and inhibitors.Chem Phys Lipids. 2016 May;197:13-24. doi: 10.1016/j.chemphyslip.2015.07.011. Epub 2015 Jul 26. Chem Phys Lipids. 2016. PMID: 26216043 Free PMC article. Review.
Cited by
-
Characterization of Temperature-Dependent Kinetics of Oculocutaneous Albinism-Causing Mutants of Tyrosinase.Int J Mol Sci. 2021 Jul 21;22(15):7771. doi: 10.3390/ijms22157771. Int J Mol Sci. 2021. PMID: 34360537 Free PMC article.
-
Non-active Site Residue in Loop L4 Alters Substrate Capture and Product Release in d-Arginine Dehydrogenase.Biochemistry. 2023 Mar 7;62(5):1070-1081. doi: 10.1021/acs.biochem.2c00697. Epub 2023 Feb 16. Biochemistry. 2023. PMID: 36795942 Free PMC article.
-
A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study.Biomolecules. 2020 Feb 4;10(2):231. doi: 10.3390/biom10020231. Biomolecules. 2020. PMID: 32033124 Free PMC article.
-
Conformational gating, dynamics and allostery in human monoacylglycerol lipase.Sci Rep. 2020 Oct 28;10(1):18531. doi: 10.1038/s41598-020-75497-5. Sci Rep. 2020. PMID: 33116203 Free PMC article.
-
Mechanistic Modeling of Monoglyceride Lipase Covalent Modification Elucidates the Role of Leaving Group Expulsion and Discriminates Inhibitors with High and Low Potency.J Chem Inf Model. 2022 Jun 13;62(11):2771-2787. doi: 10.1021/acs.jcim.2c00140. Epub 2022 May 17. J Chem Inf Model. 2022. PMID: 35580195 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

